Guest Blog from Huanming Yang on using the human genome for big-data storage
With the current annual data creation rate estimated to be in the tens of zettabytes, the flood of information currently being generated in every area of human life is crashing up against limited data storage solutions. However, DNA, which serves as a storage system for biological information, has been proposed as a potential means to store an unlimited amount of information. Here, instead of digital storage with 0’s and 1’s, the information is stored as A’s, C’s, T’s, and G’s. There have been some studies that have assessed the potential by using DNA inside living cells to store information. The problem was that the cells replicate, and introduce errors, and they also die. Getting around this problem, George Church and colleagues devised a completely in silico system, with the DNA being synthesized in short fragments on a glass chip. The authors tested the system by synthesizing DNA to contain all the information from an HTML version of a book of 53,426 words, 11 images, and one javascript file into DNA. They then read the information back by sequencing the DNA, and could restore the entire book with an error rate so low that it basically amounted to typos. While this is a great step forward, the cost of this storage system, since it requires both DNA synthesis and sequencing, still has extensive drawbacks.
 Below, in a guest blog, Professor Huanming Yang presents his thoughts on the next method that could be used to get around this issue, using human genome sequencing information as a mechanism to eliminate the need for those two steps. Prof Yang is one of the founders of BGI (our co-publisher), and has an interest in synthetic biology, previously publishing a guest blog with us on synthetic genomes.
Below, in a guest blog, Professor Huanming Yang presents his thoughts on the next method that could be used to get around this issue, using human genome sequencing information as a mechanism to eliminate the need for those two steps. Prof Yang is one of the founders of BGI (our co-publisher), and has an interest in synthetic biology, previously publishing a guest blog with us on synthetic genomes.
An in silico system for storage of information using the human genome sequence
So far, all the proposed methods or ideas for DNA information storage require the following: 1) An in silico system with relevant programs to encode and decode the information. This makes possible conversion from the information stored in a binary code (0/1 of the digital information) to then be converted to DNA sequence (A/T/C/G of the nucleotide) and vice versa. 2) A DNA synthesizer to synthesize a series of DNA nucleotides, stored either in vitro or in vivo. 3) A DNA sequencer to read the DNA sequences, and the final steps to re-convert the DNA sequence to the binary code from which it began and can be machine read. While this is a completely feasible system using current technologies, both steps 2 and 3 are very expensive and laborious, limiting the feasibility of this approach for broad use.
Here, my colleagues at BGI and I propose a fully in silico system for storing information and archiving data using DNA sequence that will be universally applicable, reliable, convenient, and inexpensive. The major advantage of our proposed system is that it eliminates the need for both DNA synthesizers and sequencers. The basic premise of the protocol is to store information in the human genome sequence.
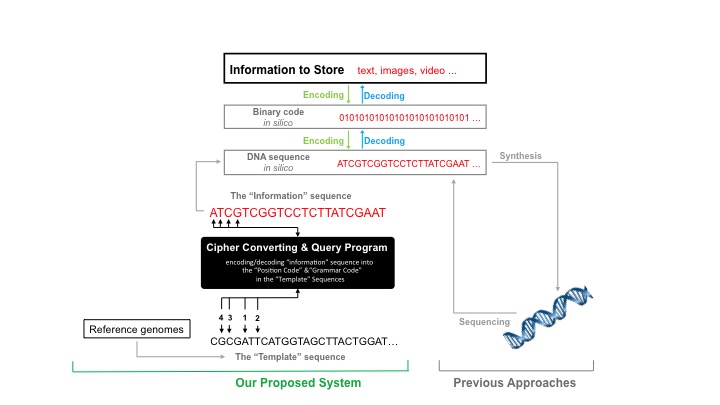
Our proposed system contains the following elements: 1) The information to store, which might be any text, images or video. These would be converted to a machine-readable binary code, followed by further conversion into DNA sequence. (This part is the same as other proposed methods or ideas) 2) A “template” sequence such as the entire human genome DNA sequence or selected regions that are without known variations, e.g. a cDNA sequence. Here, each nucleotide, A/T/C/G, in the “template” sequence is labeled with a “position code”. 3) A computer program to (a) select each required nucleotide (N=1) or a string of several consecutive nucleotides (N>1) from the “template” sequence to form the “information” sequence (Fig. 1). This is a relatively simple system that is composed of a “position code”, which indicates the position of a nucleotide in the “template” sequence, and a “grammar code”, which indicates, for example, spaces between words, beginning or end of sentences, and other grammatical components, (b) convert the “information” sequence into the binary code, and (c) reconvert the binary code back to the initial information to store.
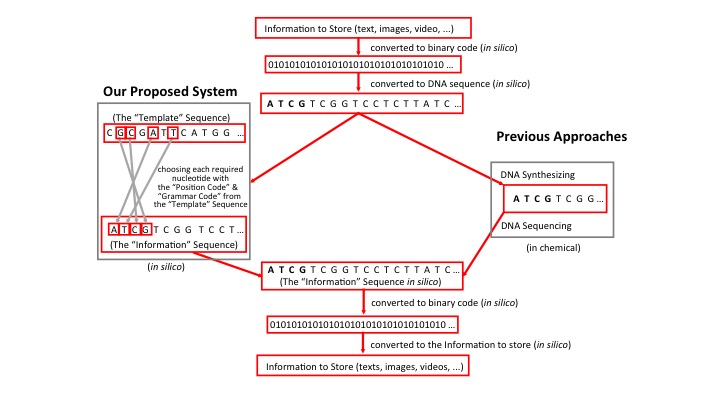 In addition, the “template” sequence could also be any other reference genome or a human-designed synthetic sequence. An “internationally standardized template sequence” could be adopted and stored in DNA molecules either in vivo as a few kb DNA insert in a vector replicating in a living organism, or in vitro as synthesized oligonucleotides in lyophilized aliquots for convenient manipulation and long-term storage.
In addition, the “template” sequence could also be any other reference genome or a human-designed synthetic sequence. An “internationally standardized template sequence” could be adopted and stored in DNA molecules either in vivo as a few kb DNA insert in a vector replicating in a living organism, or in vitro as synthesized oligonucleotides in lyophilized aliquots for convenient manipulation and long-term storage.
 It is also worth noting that this system could also be used to transfer protected or secret information at a high level of security.
It is also worth noting that this system could also be used to transfer protected or secret information at a high level of security.
In summary, this proposed system has several advantages: 1) Greatly eliminating the need to use expensive and laborious DNA synthesizers and sequencers and 2) Its capacity allows storing an unlimited amount of information. Finally, we specifically recommend use of the human genome sequence information as it is freely available to follow the HGP Spirit: “Owned by All, Done by All, and Shared by All” proposed by us and endorsed by the HGP community, and that this DNA sequence will always be present in every human being for ever and everywhere. It is fitting that storage of all of the accumulated knowledge of the human race can be stored in the in the material that encodes the information to build every human.
Feel free to add your feedback and comments via the blog commenting system, and communications should be addressed to <yanghm@genomics.cn>.
References
- Shipman SL, Nivala J, Macklis JD, Church GM: CRISPR–Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature. 2017; 547 (7663): 345-349.
- Erlich Y, and Zielinski D: DNA Fountain enables a robust and efficient storage architecture. Science. 2017; 355 (6328): 950-954.
- Grass RN, Heckel R, Puddu M, Paunescu D, Stark WJ: Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew Chem Int Ed Engl. 2015; 54 (8):2552-2555.
- Goldman N, Bertone P, Chen S, Dessimoz C, LeProust EM, Sipos B, Birney E: Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature. 2013; 494 (7435): 77-80.
- Church GM, Gao Y, and Kosuri S: Next-Generation Digital Information Storage in DNA. Science. 2012; 337 (6102): 1628.
- Cox, J. P. L: Long-term data storage in DNA. Trends Biotechnol. 2001; 19: 247–250.
Recent comment
Comments are closed.
Test comment