From Social to Biological Networks: A New Algorithm Uncovering Key Proteins in Human Disease
Published today in GigaScience is a new algorithm connecting social and biological networks to identify key proteins in Human Health. Researchers at Ben-Gurion University of the Negev have developed a machine-learning algorithm that could enhance our understanding of human biology and disease. The new method, Weighted Graph Anomalous Node Detection (WGAND), takes inspiration from social network analysis and is designed to identify proteins with significant roles in various human tissues.
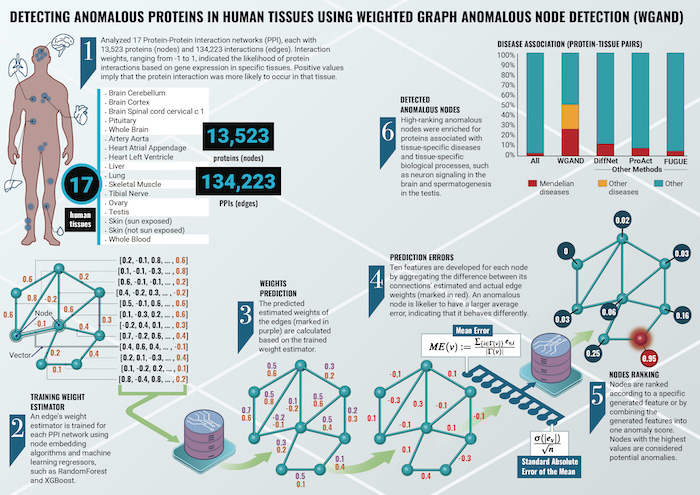
Proteins are essential molecules in our bodies, and they interact with each other in complex networks, known as protein-protein interaction (PPI) networks. Studying these networks helps scientists understand how proteins function and how they contribute to health and disease.
Prof. Esti Yeger-Lotem, Dr. Michael Fire, Dr. Jubran Juman, and Dr. Dima Kagan developed the algorithm to analyze these PPI networks to detect “anomalous” proteins—those that stand out due to their unique pattern of interactions.
The research combined the expertise in biological networks of Prof. Yeger-Lotem with the network analysis expertise of Dr. Fire derived from his study of social networks. In cybersecurity-related analyses of social networks, identifying atypical patterns can uncover fraudulent transactions or suspicious user behavior. The innovative insight of the new GigaScience study is that the same algorithms that uncover anomalies in social networks can be applied to the networks of proteins inside individual cells. By focusing on these anomalies, the algorithm can identify proteins that play crucial roles in specific tissues, such as the brain, heart, and liver.
The graph-based anomaly detection approaches adapted here previously used to detect anomalies within social networks such as identifying suspicious connections or behaviours. Or in cybersecurity detecting anomalies such as fraudulent transactions or suspicious user behavior.
WGAND successfully identified proteins associated with tissue-specific diseases, such as those involved in brain disorders and heart conditions. The algorithm also pinpointed proteins involved in critical biological processes, like neuron signaling in the brain and muscle contraction in the heart. Moreover, WGAND outperformed other existing methods in terms of accuracy and precision.
“This innovative algorithm has the potential to pinpoint which proteins are important in specific contexts, helping scientists to develop more targeted and effective treatments for various conditions,” says Prof. Yeger-Lotem.
“It’s exciting to see how bringing together expertise from bioinformatics and cybersecurity can lead to breakthroughs in understanding human biology. By applying network analysis and machine learning, we’ve developed a tool that helps uncover key proteins in different tissues—paving the way for new insights into human health and disease,” says Dr. Michael Fire.
Regular readers will have seen us publishing other examples of how Dr. Fire has applied AI approaches to many different fields, from social media and bibliometric network analysis for many different applications, from predicting infectious disease outbreaks to the over-optimization of academic publishing metrics.
In accordance with Dr. Fire’s principles as the head of the Fire AI Lab, the WGAND algorithm is open-source, allowing researchers worldwide to utilize and build upon it. The Yeger-Lotem lab also maintains web tools enabling easy access to researchers with no computational background.
This work is an interesting case study on how the merging of expertise in bioinformatics and cybersecurity can lead to significant insights into complex biological questions. Exemplifying the power of interdisciplinary collaboration in fueling scientific breakthroughs. The intersection of diverse fields such as bioinformatics, machine learning, and network analysis fosters innovation, driving advancements that can swiftly translate into practical applications in healthcare. To provide additional background insight into the paper, and also discuss how such interdisciplinary collaborations can get off the ground, we organised a Cassyni author webinar with Prof. Esti Yeger-Lotem and Dr. Michael Fire, and the record of this is now available here.
References
Kagan D, Jubran J, Yeger-Lotem E, Fire M. Network-based anomaly detection algorithm reveals proteins with major roles in human tissues. GigaScience. 2025 Jan 6;14:giaf034. doi: 10.1093/gigascience/giaf034
Fire M, Guestrin C. Over-optimization of academic publishing metrics: observing Goodhart’s Law in action. GigaScience. 2019 Jun 1;8(6):giz053. doi: 10.1093/gigascience/giz053.
Kagan D, Moran-Gilad J, Fire M. Scientometric trends for coronaviruses and other emerging viral infections. GigaScience. 2020 Aug 1;9(8):giaa085. doi: 10.1093/gigascience/giaa085.
. Cassyni Webinar. https://doi.org/10.52843/cassyni.06hwvh