A neuroinformagical reunion at the virtual INCF Assembly
The International Neuroinformatics Coordinating Facility (INCF) hosted their Neuroinformatics Assembly on 19th-23rd April 2021. This conference aims to highlight open innovation in neuroscience, and additionally to showcase recently developed software tools and data infrastructures. Being a community representing the most forward thinking and open science friendly members of the neuroscience community, we’ve been long term attendees and supporters of the meeting (see GigaBlog write-ups of the 2015 and 2019 meetings). With the pandemic still preventing in-person meetings, it was great to have a virtual catch-up with lots of our friends and Editorial Board members, and it was great to see the number of our authors presenting this year too. This year GigaScience Data Scientist Chris Armit attended the virtual INCF conference and reports below on some of his major highlights.

The Importance of Data Sharing
As a neuroimaging researcher, in her talk Elizabeth DuPré (McGill University) summed up essentially one of the core tenets of INCF by stating, “openly sharing data creates new opportunities in scientific tool development (and) research”. Elizabeth followed this up with a use case study – available as a preprint on bioRxiv – that shows extensive distributions of neuroimaging data points (Figure 1). These distributions become quite apparent when analysing collections of brain images from multiple sources, and a fundamental message of Elizabeth’s recent analysis is that, if a researcher were restricted to looking at in-house data, these distributions would potentially go unnoticed. Of note, Elizabeth and colleagues have recently published in GigaScience a comparative performance evaluation of preprocessing pipelines, which partially explains the source of inconsistencies in neuroimaging findings. As Elizabeth points out in her talk, “benchmarking across multiple datasets provides more reasonable estimates of performance”.
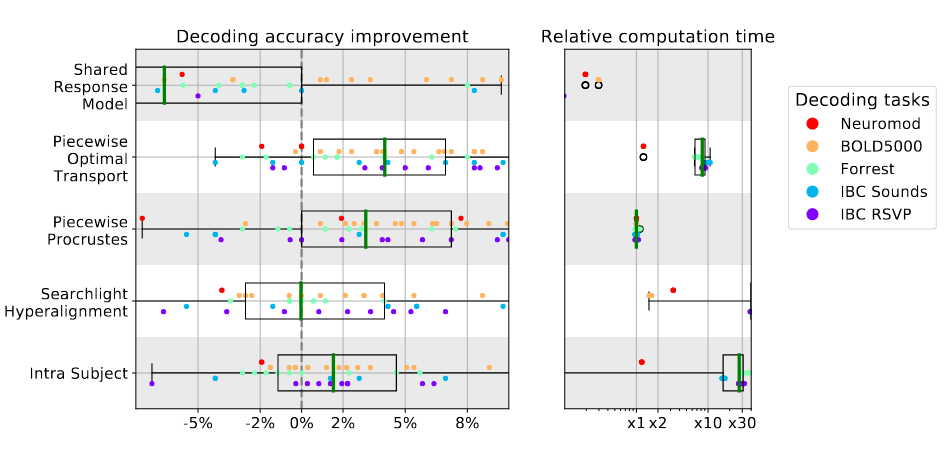
Figure 1 – Decoding accuracy improvement and computation time after whole-brain functional alignment. Source: bioRxiv https://doi.org/10.1101/2020.12.07.415000
Finding Neuroinformatics Tools and Resources
So how would a researcher find and run these neuroimaging pipelines? In his talk, Tristan Glatard (Concordia University) reported on the Canadian Open Neuroscience Platform (CONP), which enables researchers to find neuroimaging pipelines easily, and ensures that they are made available as container images – such as those of Docker, Singularity, or rootfs formats – that enable reuse with relatively simple and straightforward installation instructions. These pipelines can also be run locally using Boutiques – published in GigaScience – which is a system to “automatically publish, integrate, and execute command-line applications across computational platforms” (Figure 2). A core concept here is ‘FAIRness’ – ensuring tools and resources are Findable, Accessible, Interoperable, and Reusable. From a reuse perspective, the CONP infrastructure and the Boutiques framework are incredibly useful for the entire neuroinformatics community.
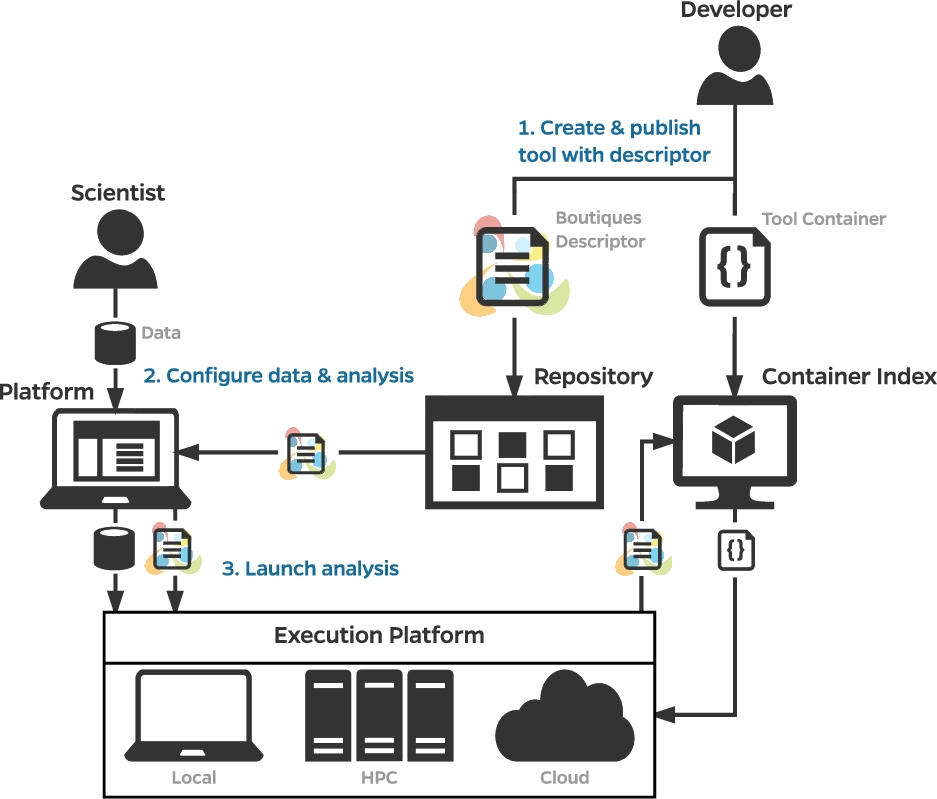
Figure 2 – Publication, integration, and execution of applications with Boutiques. Source: GigaScience https://doi.org/10.1093/gigascience/giy016
In a similar vein, Hervé Menager (Institut Pasteur) reported on bio.tools, which is a comprehensive registry of software and databases that enables biomedical researchers “to find, understand, utilise and cite the resources they need in their day-to-day work”. biotoolsSchema – the formalized XML schema used for bioinformatics software description – has recently been published in GigaScience and “promotes the FAIRness of research software, a key element of open and reproducible developments for data-intensive sciences” (Figure 3). bio.tools provides a very useful portal for researchers who are unfamiliar with the suite of publicly available neuroinformatics tools and resources that are available for reuse. Hervé additionally detailed how bio.tools helps software developers. Indeed, as Hervé explains, when you go to register your software in bio.tools, you may find that your tool is already there – a very helpful feature that ensures rapid dissemination of software tools and resources – in which case you simply have to claim your software.
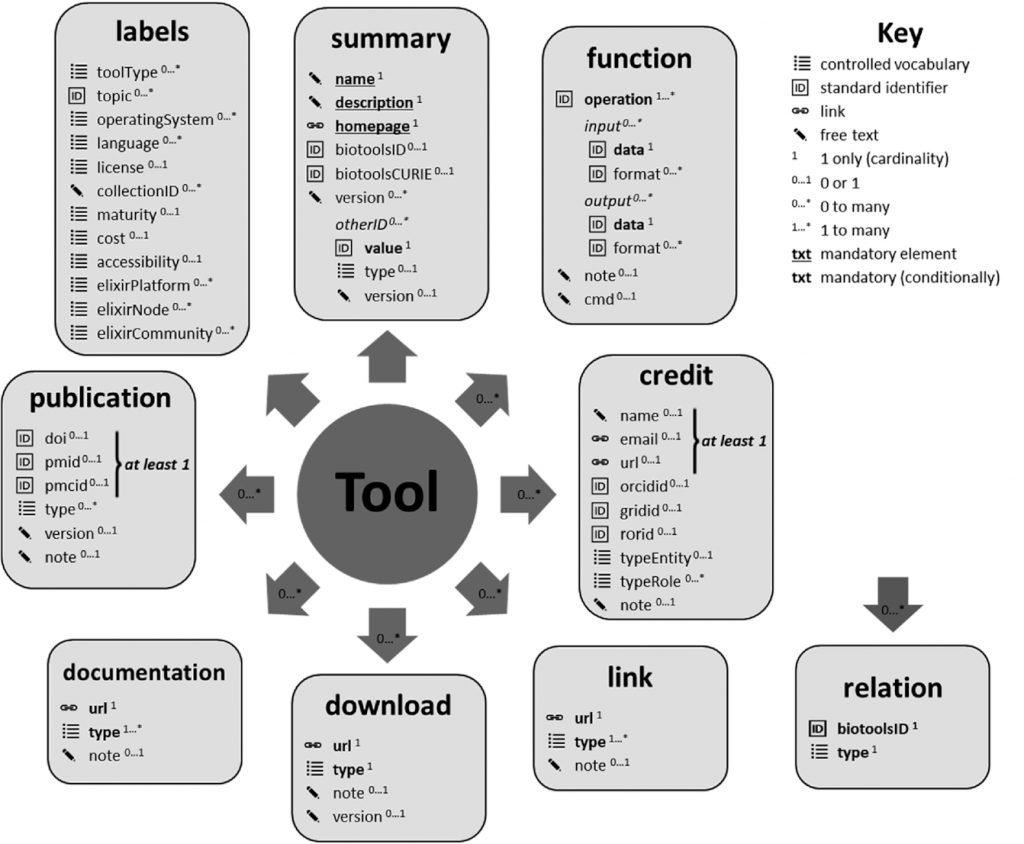
Figure 3 – biotoolsSchema overview. Software attributes are organized into 9 groups (in boxes) and include terms from controlled vocabularies defined internally within biotoolsSchema, standard identifiers (including from the EDAM ontology), links, or free text. source: GigaScience https://doi.org/10.1093/gigascience/giaa157
Neuroinformatics Standards and FAIRsharing
There was great emphasis at this year’s meeting on the standards that are most relevant for the neuroinformatics community. Standards are necessary to ensure that software tools and data are interoperable, and they provide the necessary list of requirements to ensure interoperability between different labs. BIDS (Brain Imaging Data Structure) was arguably the most widely accepted standard for this community. Sylvain Takerkart (CNRS-AMU) described BIDS as “a simple and intuitive way to describe neural data” and highlighted that BIDS is now used as a standard for MRI, MEG, EEG, and iEEG data. GigaScience Editorial Board Member Susanna-Assunta Sansone further presented FAIRsharing, which is a web-based searchable portal of interlinked registries of standards, databases and data policies, and which provides “curated, community-vetted descriptions and knowledge graphs that represent these resources and their inter-relationships” (see GigaBlog on FAIRsharing). Scholarly publishers – including GigaScience – use FAIRsharing to confidently guide researchers to resources that are recommended in their data policy. INCF have additionally created a FAIRsharing collection of endorsed standards – tailored to the needs of the neuroinformatics community – and this lists BIDS, NeuroData without Borders, NeuroML, Neuroscience Information Exchange Format, and PyNN as the top five recommended standards for this community.
Open Tools for Electrophysiology and Connectomics
Of the various tools that were showcased at the conference, I was most intrigued by Elephant – an open-source Electrophysiology Analysis Toolkit – and The Virtual Brain (TVB), a neuroinformatics platform that utilises connectivity data for full brain network simulations (Figure 4). Sylvain Takerkart highlighted the utility of electrophysiology analysis tools, such as Elephant, which can provide statistics of spike trains, but also more advanced statistics such as Unitary Event Analysis, which detect coordinated spiking activity in the brain. Petra Ritter (Berlin Institute of Health) presented The Virtual Brain (TVB), which is a fabulous resource that simulates brain behavior based on findings in clinical scenarios, such as what would be obtained from EEG, MEG, and fMRI scanners. A core concept here is that the simulation is simpler than the human brain, but has sufficient complexity to render a model that is nevertheless realistic. Petra addressed this point in her talk, and quotes the eminent Austrian neurologist and psychiatrist Victor Frankl in stating, “a good theoretical model of a complex system should be like a good caricature: it should emphasise those features which are most important and should downplay the inessential details”. The Virtual Brain (TVB) web portal provides integrated simulation tools, pipelines and datasets for the neuroinformatics community.
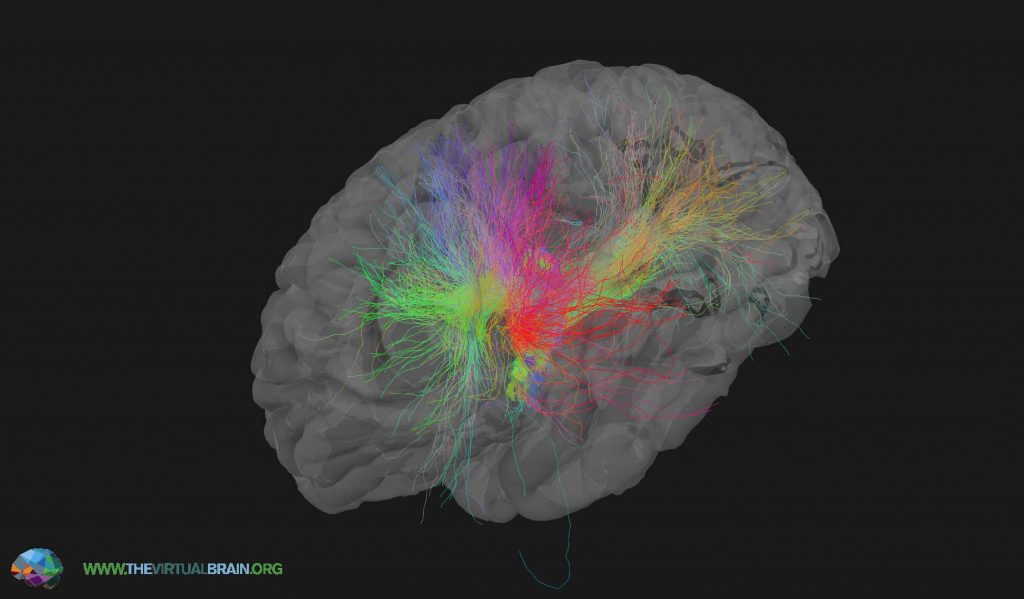
Figure 4 – The Virtual Brain delivers simulations of connetomic data analysis. Source: https://www.thevirtualbrain.org/tvb/zwei/image-zoom/166699/3?stage=brainsimulator
Deep Learning and Behaviour Analysis
In the session on “Data science and neuroinformatics” Mackenzie Mathis (Swiss Federal Institute of Technology Lausanne) highlighted the utility of the software tool DeepLabCut, which is a remarkable toolbox for efficient markerless pose estimation. As Mackenzie explains, in behavioural analysis a key objective is to use computer vision to capture the essence of behaviour, and pose estimation is one of the major computational challenges. To highlight the issue, Mackenzie first showcased marker-based pose estimation, where for example an actor wears a suit with reflectors and this is used to capture the movement of the actor. This computational facsimile can then be used to drive CGI animations that correspond to the actor’s movements. A key component of this scenario is that there is a priori knowledge of where the markers are placed. In contrast, in markerless pose estimation, transfer learning with deep neural networks enables automated tracking of predicted keypoints in a time-series of images, and this enables generation of a highly accurate post-hoc skeleton (Movie 1). Remarkably, in the example shown of the movement of the mouse forepaw, only 141 training images were used. This is a powerful software tool with huge potential for investigations of brain and behaviour. As Mackenzie explains, “pose estimation can help us answer new questions in motor control.” For more enticing use case examples of this software tool, see DeepLabCut Model Zoo. DeepLabCut is openly available on GitHub where it has been assigned an Open Source Initiative-approved LGPL-3.0 license.
Neuroinformatics and Social Change
Perhaps the most thought-provoking talk of the conference was Pedro Valdés-Sosa (Cuban Neuroscience Center) who offered a series of vignettes on the importance of neuroinformatics to our understanding of health and disease. The most poignant of these was Pedro’s discussion of a published study of an EEG Source Classifier that could identify children malnourished in the first year of life from the EEG abnormalities that are observed 45+ years later. The observed overall loss of neural activity in individuals with a history of Protein Energy Malnutrition, with 0.82% accuracy relative to healthy controls, suggests a delay in brain development in malnourished children and highlights the potential of neuroinformatics in a societal context. One hopes that these research findings can be used to inform policy change and good governance in tackling issues such as childhood malnutrition in the Global South, and the INCF are to be commended for taking the first bold steps towards confronting these challenges.
You can watch the virtual INCF assembly talks in the recorded live stream on the INCF vimeo channel, which is another advantage of a virtual meeting such as this. We look forward to the next meeting and further neuroinformagical catch-ups in the future.