Sex chromosome madness in the iconic echidna
This week we published a new, high-quality genome sequence of one of Australia’s most iconic animals, the short beaked echidna. The almost gapless genome sequence of this egg-laying mammal helps researchers to track genomic reshuffling events that gave rise to a perplexing sex determination system.
At first glance they may be mistaken for a weird-looking hedgehog. However, the short-beaked echidna (Tachyglossus aculeatus) belongs to a unique group of egg-laying mammals called “monotremes” (with the platypus as the other prominent member). The international team of authors that embarked on this project include Guojie Zhang and Qi Zhou at the Centre for Evolutionary & Organismal Biology at Zhejiang University, Yang Zhou from BGI-Research and Frank Grutzner from Adelaide University. The work is part of the international Vertebrate Genome Project, and hosted in the VGP Genome Ark Database.

The short-beaked echidna (Tachyglossus aculeatus)
Among other unusual features (egg-laying mammals – helllooo???), the unique position of monotremes in the tree of life is also reflected in their genomes, and in particular in the way sex is determined genetically. In most sex determination systems, sex chromosomes typically come as a pair, such as in the familiar X/Y system in humans and in most other mammals. In this familiar setup, sperm carry either an X or a Y chromosome. Together with the X chromosome that the egg contributes, females typically have two X chromosomes, and males one X and one Y.
Sex chromosomes galore
In contrast, the situation is far more complex in monotremes, featuring multiple X and Y chromosomes. As if this was not complicated enough already, there are also differences between the two monotreme groups, differentiating echidnas and the platypus. In male echidna there are nine sex chromosomes, in platypus even ten (five X and five Y in male platypus, and five X but only four Y chromosomes in male echidna). How on earth is genetic disaster avoided during meiosis, when sex chromosomes need to be passed on to egg and sperm cells? The trick is that these sex chromosomes pair in a head-to-tail manner and form chains. This allows for them to be distributed in an orderly manner to the germ cells.
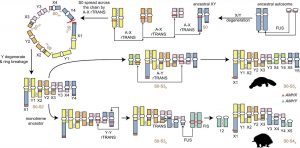
Model for the sex chromosome evolution in monotremes. (from Zhou et al, CC-BY)
The details of this process, and the intriguing question of how and why this complicated system originated in the first place, is still under investigation. The new echidna genome sequence presented in GigaScience helps to trace the genetic events that led to this remarkable chromosomal arrangement. A previous version of an echidna genome has been published before, but the new high quality sequence with very few gaps allows researchers to infer events like chromosomal rearrangements in the ancestry line of monotremes.
Zhang says:
“The high quality reference genome of echidna allows us to infer which chromosomes are shared between echidna and platypus since they diverged from other mammals and which were newly evolved more recently after the two monotreme split”.
The origins of the echidna sex chromosomes
The evolutionary lineages of echidnas and platypus have diverged around 55 million years ago and there is evidence that chromosome fusion or fission events might have occurred since the divergence of platypus and echidna. Using both the new echidna and an existing platypus genome sequence, the GigaScience authors reconstructed the likely chromosome composition of the monotreme ancestor, using also chromosomal assemblies of placentals (human, bovine and sloth), marsupials (opossum and Tasmanian devil), and reptilian outgroups (chicken, turtle and common wall lizard).
Analysis of the sex chromosomes revealed that the ancestral X chromosome of monotremes had exchanged segments with ancestral non-sex chromosomes (autosomes), which gave rise to the complex system.
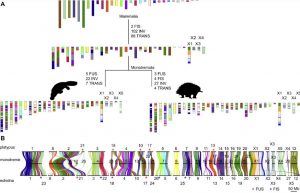
Karyotype evolution in monotremes. Figure 2 of Zhou et al (cc-BY)
Gene family expansion
In addition to these structural changes in genome organisation, the authors also found signals of gene family expansion, meaning that additional copies of specific genes arose during the evolution of monotremes. One such event concerns a gene called SYCP3Y. This gene encodes a component of the synaptonemal complex, which is a protein structure that connects chromosome pairs during meiosis. SYCP3Y is duplicated from autosomes and expanded in both platypus and echidna, suggesting that the gene expansion may be associated with the evolution of the sex chromosome complex or play a role in its organization.
Overall, the new results provide a detailed evolutionary history of the highly unusual multiple sex chromosome system in monotremes and demonstrate how today’s high-resolution genomic methods can uncover evolutionary events that shaped the mammalian tree of life millions of years ago.
Share the press release:
https://www.eurekalert.org/news-releases/1069499
References:
Zhou Y et al., Chromosome-level echidna genome illuminates evolution of multiple-sex-chromosome system in monotremes. GigaScience. 2025. https://doi.org/10.1093/gigascience/giae112
Data in GigaDB: Zhou Y, Jin J, Li X, et al. Supporting data for “Chromosome-Level Echidna Genome Illuminates Evolution of Multiple Sex Chromosome System in Monotremes.”. GigaScience Database 2024. https://doi.org/10.5524/102609
Rhie, A., McCarthy, S.A., Fedrigo, O. et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 (2021). https://doi.org/10.1038/s41586-021-03451-0
Kerstin Howe, William Chow, Joanna Collins, Sarah Pelan, Damon-Lee Pointon, Ying Sims, James Torrance, Alan Tracey, Jonathan Wood, Significantly improving the quality of genome assemblies through curation, GigaScience, Volume 10, Issue 1, January 2021, giaa153, https://doi.org/10.1093/gigascience/giaa153
Sheri Sanders. DNA Day 2019: How to sequence the genomes of the weird and wonderful. In GigaBlog. https://doi.org/10.59350/7×637-mb215