Meet the GigaScience ICG Prize Winners, Pt. 1: Aequatus
 This is the Dawning of the Age of Aequatus
This is the Dawning of the Age of Aequatus
Our ICG Prize is over now for another year, and we’ll shortly follow up with an announcement on which of the six winners won the $1000 first prize. To help you see how great all the entries were we will introduce and profile some of the winning papers. With the accompanying articles now passing through peer review and highlighted in our series page, the most recent winning paper published is “Aequatus: An open-source homology browser”. First author Anil Thanki came out to Shenzhen to present the work, and if you didn’t make ICG the video of the talk is embedded below, and his slides are also included here.
Aequatus is a new bioinformatics tool developed at the Earlham Institute (formerly TGAC) in Norwich, helping to give an in-depth view of syntenic information between different species, providing a system to better identify important, positively-selected, and evolutionarily-conserved regions of DNA.
Generally, organisms that are closely related show a high degree of synteny i.e. they possess similar sequences along their chromosomes, where closely related genes that are presumed to have the same function are clustered in a similar organisation between species. Thus, many human genes have high synteny with mammals, from chimpanzees to mice.
Studying the synteny between organisms can help us to identify how genetic regions change through evolution, and has far-reaching applications – including better understanding evolution and how we came to be, aiding studies into human health, as well as in breeding better crops.
 Of the utility of the tool Anil Thanki says: “We are very excited about Aequatus because it provides a really intuitive way to visualise homologous genes among species. Aequatus provides a seamless user experience using the latest web technologies available to represent genomics data. It helps biologists delve into the details of homologous genes by comparing them at the genomic feature level. We have also connected this resource with the SMART protein domain information server to let researchers get to relevant data without having to switch services.
Of the utility of the tool Anil Thanki says: “We are very excited about Aequatus because it provides a really intuitive way to visualise homologous genes among species. Aequatus provides a seamless user experience using the latest web technologies available to represent genomics data. It helps biologists delve into the details of homologous genes by comparing them at the genomic feature level. We have also connected this resource with the SMART protein domain information server to let researchers get to relevant data without having to switch services.
Built using open-source technologies (a requirement for GigaScience), Aequatus provides a fast and intuitive web-based browsing experience to bridge the gap between phylogenetic changes and gene feature information.
The development of Aequatus gave rise to an open-source JavaScript library – Aequatus.js – which retains the functions of the full visualisation application but can be integrated with other web applications, such as the Galaxy workflow system (a favourite platform of ours, with our growing Galaxy series a testament to this).
One such application is the also published in GigaScience GeneSeqToFamily tool, a Galaxy workflow based on the Ensembl Compara GeneTrees pipeline to find gene families. The Aequatus plugin has been made available within Galaxy (currently on usegalaxy.eu) in order to visualise resulting gene families garnered from GeneSeqToFamily.
Whereas traditional phylogenetic trees (a visualisation of the shared ancestry in a “family tree”) present an overview of synteny, Aequatus also provides information regarding structural changes in genes, including variation within them that corresponds to changes in phenotype (appearance).
A novel, more complete visualisation tool
Using a “guide” gene as a reference, other genes are mapped based on alignment (an analysis of sequence similarity, or how closely two genes are related to each other based on their DNA or protein sequence). Alignments are retrieved from open-source databases, Ensembl Compara and the Ensembl Core, then Aequatus processes both comparative and feature data to provide a visual representation of phylogenetic and structural changes between species based on a shared colour scheme.
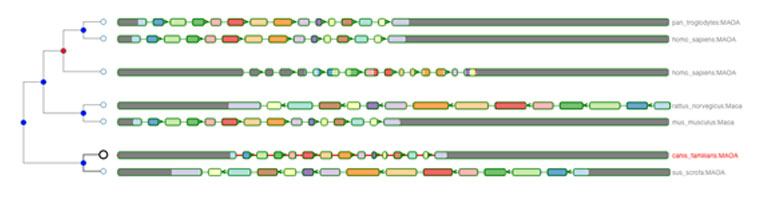
A typical gene tree visualised using the Aequatus tool.
This helps to visualise regions of homology, while also allowing the identification of changes to genes, such as insertions or deletions, with black bars representing insertions specific to a given gene compared to the “guide”. Overall, Aequatus provides a unique way to explore complex relationships between genes from various species at a level that has so far been unrealised. Applicable not only to high-quality reference genomes including mouse and human, Aequatus has been designed for use with hard-to-assemble or non-model organisms. The latest version of Aequatus also supports the Ensembl REST API, which can retrieve data directly from Ensembl server and doesn’t necessitate the use of local data improving the portability of Aequatus.
Senior author Rob Davey said: “It’s great to see this work published and indeed selected for an award at an international conference. This shows that visualisation of genomic data is still an active and valuable area of research. Aequatus can really help researchers gain access to even more fine-grained information about their genes and organisms of interest.”
Further Reading
Thanki AS, Soranzo N, Herrero J, Haerty W, Davey RP. Aequatus: An open-source homology browser. Gigascience. 2018 Nov 5. doi: 10.1093/gigascience/giy128.
Thanki AS, Soranzo N, Haerty W, Davey RP. GeneSeqToFamily: a Galaxy workflow to find gene families based on the Ensembl Compara GeneTrees pipeline. Gigascience. 2018 Mar 1;7(3):1-10. doi: 10.1093/gigascience/giy005.
This post was adapted with kind permission from the Earlham Institute, and please use access Aequatus at: http://www.earlham.ac.uk/aequatus