GigaByte and River Valley Technologies push the boundaries of Executable Research Articles using Stencila and Code Ocean
GigaScience Press and River Valley Technologies, with the help of Stencila, launch their first interactive “Executable Research Article” from their new scientific journal, GigaByte. With a new immunoinformatics tool combined with interactive examples applied to the study of coronavirus immunity, this new publication underlines the importance of speed, accuracy and trust in scientific communication brought to light by the COVID-19 pandemic.
Today GigaByte publishes its first Executable Research Article (ERA), using technology from Stencila and Code Ocean to showcase interactive and executable versions of the figures. This ERA allows readers to use two different platforms to inspect the code, to modify it, and then to re-execute it directly within the article. This is a transformative step in scientific publishing as it changes an article from a static object into a living document, thus not only improving reproducibility, but making the article directly reusable by the reader. GigaScience Press has experimented before with Galaxy publications in a custom GigaGalaxy server, as well as Code Ocean deployment of executable code-bundles, but this is the first time working with the customisability and interactivity provided by the ERA-format and technology provided by Stencila.
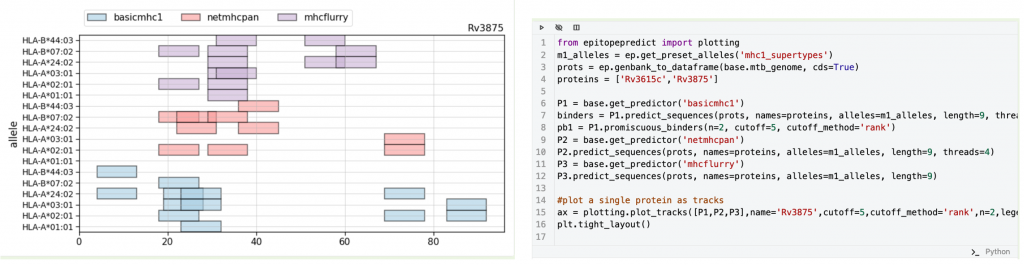
This technology is showcased in a “Technical Release” article, which is designed to present and credit the release of software and computational pipelines. The article presents a new immunoinformatics tool, epitopepredict that predicts likely protein sequences involved in the adaptive immune response, providing computational insight for vaccine design and immuno-diagnostics prior to carrying out expensive laboratory experiments.
Epitopepredict has many applications and is particularly timely in the COVID-19 pandemic, so this article serves as a well timed first ERA in GigaByte. Specifically, it is of considerable interest to determine whether other components of the immune system, in addition to antibodies, are important in protecting individuals. The work described in the GigaByte ERA showed that epitopepredict can be used to predict whether memory T cells generated from previous exposure to the human common cold coronaviruses are potentially cross-reactive against the SARS-CoV-2 coronavirus that causes COVID-19.
In order to make the article reproducible, Stencila used their open source software to convert the author’s word processor file to a Jupyter Notebook, worked with the author to add code, and then converted the article to a highly semantic, machine readable and interactive HTML version. Stencila even created a GigaByte theme for this interactive version, demonstrating the flexibility and scalability of their platform.
Nokome Bentley of Stencila is excited by the next steps in the evolution of ERAs: “We were really pleased with how we could easily and quickly apply our format conversion tool and theming framework to rapidly deliver this ERA for GigaByte. We are looking forward to creating an even tighter integration with River Valley’s XML-based publishing stack to remove the need to have a parallel version and instead embed interactive web components in the main article”.
Beyond using Stencila to create an ERA, the journal integrated and presented individual figures, the data, code, and computation environment in a Code Ocean “Compute Capsule”. This allows readers to directly interact with an embedded version of the Code Ocean platform in the article; additionally, readers can immediately deploy and run this in their own cloud computing account (see embedded below).
Author Damien Farrell says of the process putting the reproducible article together: “Jupyter notebooks are a core part of my workflow. Stencila was easy to integrate with my notebooks and with minimal changes allowed them to be turned into dynamically generated articles.”
Looking toward the future, the University College Dublin researcher commented on the overall benefits of this new approach, saying: “I hope using Code Ocean and Stencila will show the benefits of using an open source model (and Python!). Reproducibility is still a major challenge in computational biology as in other fields. These new publishing tools being used by GigaByte are going to become increasingly important as we move away from the old publishing models.”
Kaveh Bazargan of River Valley Technologies, who has been keenly interested in and committed to adding the ability to publish ERAs as a central component of River Valley Technologies’ publishing platform, highlighted how eLife made this possible, stating: “The Executable Research Article format was conceived by eLife‘s collaboration with Stencila and developed as an open source technology. GigaByte is the second journal to use this technology. We all acknowledge eLife for helping seed this new ecosystem for disseminating research.”
You can test out the interactive version of the manuscript here: https://gigabyte.stencila.io/epitopepredict/
To learn more about ERA, visit: https://elifesci.org/era
This is GigaByte‘s first example of a Technical Release article, and if you have software tools and pipelines to share in an open and reproducible manner please contact us or submit now as currently there are no APCs.
Further Reading:
Damien Farrell. epitopepredict: A tool for integrated MHC binding prediction. GigaByte, 2, 2021 https://doi.org/10.46471/gigabyte.13
UPDATE 02/03/2020: eLife and Stencila has just announced the next phase of development for the ERA project, and you can read more about it in this posting (including a nice mention of our integration too).