Rewarding Reproducibility: First Papers in our Galaxy Series utilizing our GigaGalaxy platform
Push the button! GigaScience moves toward more interactive articles

Research articles are being published with increasingly large and complicated supporting datasets, together with the software code used in analyses of the data. However, there is a growing number of studies reporting the inability to reproduce previously published findings which may, at least in part, be responsible for the increasing rate of retractions that Bjorn Brembs has calculated will overtake the number of papers published some time in the mid-2040s. Furthermore, there is an awareness of the “reproducibility gap” within the scientific community, with Francis Collins of the NIH just publishing a statement expressing concern about this issue. Whilst this has provoked some to deny this is a serious issue (ironically in journals with the highest retraction rates), GigaScience has joined initiatives such as Sage Bionetworks Synapse and Science Exchange reproducibility initiative in attempting to do something about this through building platforms and procedures to assist and reward authors who are keen on their work being reproducible and actually used. The Galaxy community is one that shares similar goals, with a computational platform which allows users to share workflows, histories and wrapped tools in an easy-to-use and open source interface that even people without coding experience can use. On top of our GigaDB repository to host large scale datasets, we have also set up our own Galaxy server called GigaGalaxy to similarly present and host the computational outputs and methods of studies published in GigaScience.
Attending the Galaxy Community Conference in Oslo last summer (see the write-up here), we and the conference committee announced a call for papers for a special thematic focused series on studies utilizing large-scale datasets and workflows. The initial results of this are now available, with the first two papers just out and available from the new series page. Whilst the series considers best practice papers, discussion, as well as novel uses of Galaxy, these first papers are examples of Galaxy toolkits, with a genome diversity tool collection presented from the Webb Miller lab at Pennsylvania State, and a set of analytical and visualisation tools for Complete Genomics sequencing data from the Stubbs lab at Erasmus Medical College. What differentiates this series from other traditional journals are doing is the focus on reproducibility, and the use of permanent DOIs and our own Galaxy server that can archive and present wrapped tools and workflows and histories from the papers.
DOIs for workflows
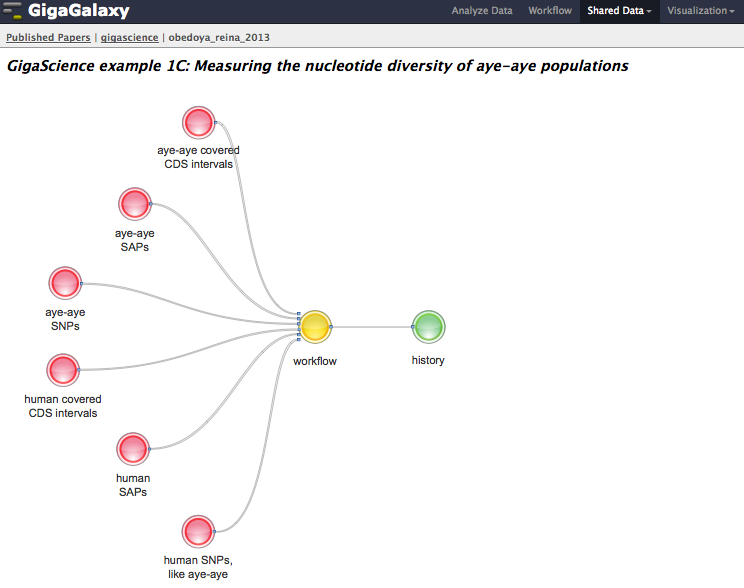
Following on from our experiences allocating DOIs to software from papers, we helped lobby DataCite to include “workflow” as a similar resource type, and they have now included this in the release of their latest metadata schema. We have been testing our GigaGalaxy platform by implementing workflows from our SOAPdenovo2 publication, and we presented much of this work at our “What Bioinformaticians need to know Beyond the PDF” workshop at ISMB (which incidentally will be continued again at the 2014 meeting). The genome diversity toolkit is our first example of a DOI that resolves purely to a workflow. From the landing page in GigaDB, you can download the Galaxy XML files, or click the link to appropriate part in the “Papers” section of our GigaGalaxy server to browse and run the workflows discussed in the paper. Presenting a handy toolkit covering a number of popular population genetics tools, the paper provides diverse examples of their application on the genetics of Lemurs, and Canines and Cave Bears (oh my!). Seven of these examples are viewable on our GigaGalaxy page. Whilst the latest versions of the tools and further examples are available from the main Galaxy server and the authors website, the GigaScience Galaxy server provides access to static versions of the tools used within the examples of the paper. To make it a little more interactive and understandable for users, we have produced SVG graphs to help visualize how input datasets, workflows and histories are related to each example analysis.
As a step on the road to executable papers, this is aiming to be a more interactive and two-way experience than traditional publication models still rooted in the print era, so please feedback to us at editorial@gigasciencejournal.com on on any bugs or features you would like to see. We would like to thank our collaborators at CBIIT (the CUHK-BGI Innovation Institute of Trans-omics) who helped us set up our Galaxy server, BioMed Central, DataCite, as well as the authors and reviewers for working with us to get these examples out and online. The series is still open and we are continuing to take and review submissions, so watch the series page for future additions. Please contact us if you are interested in submitting your work or submit through our submission system here. Thanks to support from the BGI our article processing charges are still currently free and, as we are again silver sponsors of the 2014 Galaxy Community Conference, we hope to meet many of you there.
References
1. Bedoya-Reina et al.: Galaxy tools to study genome diversity. GigaScience 2:17 http://dx.doi.org/10.1186/2047-217X-2-17
2. Hiltemann et al.: CGtag: complete genomics toolkit and annotation in a cloud-based Galaxy. GigaScience 2014 3:1 http://dx.doi.org/10.1186/2047-217X-3-1
3. Bedoya-Reina, OC; Ratan, A; Burhans, R; Kim, HL; Giardine, B; Riemer, C; Li, Q; Olson, TL; Loughran Jr, TP; vonHoldt, BM; Perry, GH; Schuster, SC; Miller, W (2013): GigaGalaxy workflows and histories from “Galaxy tools to study genome diversity” GigaScience Database. http://dx.doi.org/10.5524/100069
4. GigaScience Galaxy Series Page https://academic.oup.com/gigascience/pages/galaxy_series_data_intensive_reproducible_research
Recent comments
Comments are closed.
[…] Push the button! GigaScience moves toward more interactive articles Research articles are being published with increasingly large and complicated supporting datasets, together with the software code used in analyses of the data. However, there is a growing number of studies reporting the inability to reproduce previously published findings which may, at least in part, be responsible for the increasing rate of retractions that Bjorn Brembs has calculated will overtake the number of papers published some time in the mid-2040s. Furthermore, there is an awareness of the “reproducibility gap” within the scientific community, with Francis Collins of the NIH […]
[…] help visualize the datasets, workflows, and histories in a publication, and then execute them. Read their blog post for […]
[…] and executed through our GigaGalaxy server (see our recent posting on this), but on top of integrating workflows into our papers through citable DOIs, the papers themselves can be generated (and subsequently reproduced) in a similar manner using a […]
[…] our goal to publish different shaped research objects to the traditional static paper, on top of publishing Galaxy workflows and packages to recreate papers in Knitr, this new paper provides another great example of a more […]
[…] clearer visualisation of results in our papers (see more on its functionality and our Galaxy series in GigaBlog. We’ve just published our third paper in the series and have another one in […]