GigaScience at ESRIC Super-Resolution Clinic
The diffraction limit of a microscope hinders the ability to see single molecules as the optics do not allow the researcher to distinguish between two fluorescently labelled molecules that are less than 200nm apart. As a means of overcoming this barrier, super-resolution microscopy utilises various tricks to go beyond the diffraction limit and image sub-cellular nanostructure in cells and tissues. This was the theme of the ESRIC Super-Resolution Clinic in Heriot-Watt University, Edinburgh on 28th Feb 2019. GigaScience Data Scientist Chris Armit was there and was deeply impressed with the panoply of emerging technologies that are available at this world-class imaging facility.
As ESRIC manager at Heriot-Watt Campus Ali Dun explained, super-resolution imaging exploits various techniques, some of which are more suited to particular experiments. To illustrate with one example, Structured Illumination Microscopy (SIM) utilises the Moiré effect that is formed when periodic light patterns are rotated, and this technique allows cells and tissues to be imaged at approximately 100nm resolution, which is twice the resolution of the diffraction limit. A key advantage of this imaging method is that it utilises the standard fluorophores that researchers would use in confocal imaging. To obtain single-molecule resolution, researchers can use techniques such as Stochastic Optical Reconstruction Microscopy (STORM) that utilise photoswitchable fluorophores that blink. By monitoring the blinking fluorophores over time, researchers can obtain incredible spatial resolution of 20nm in the lateral dimension that enables single-molecules in living cells to be identified and followed through time.
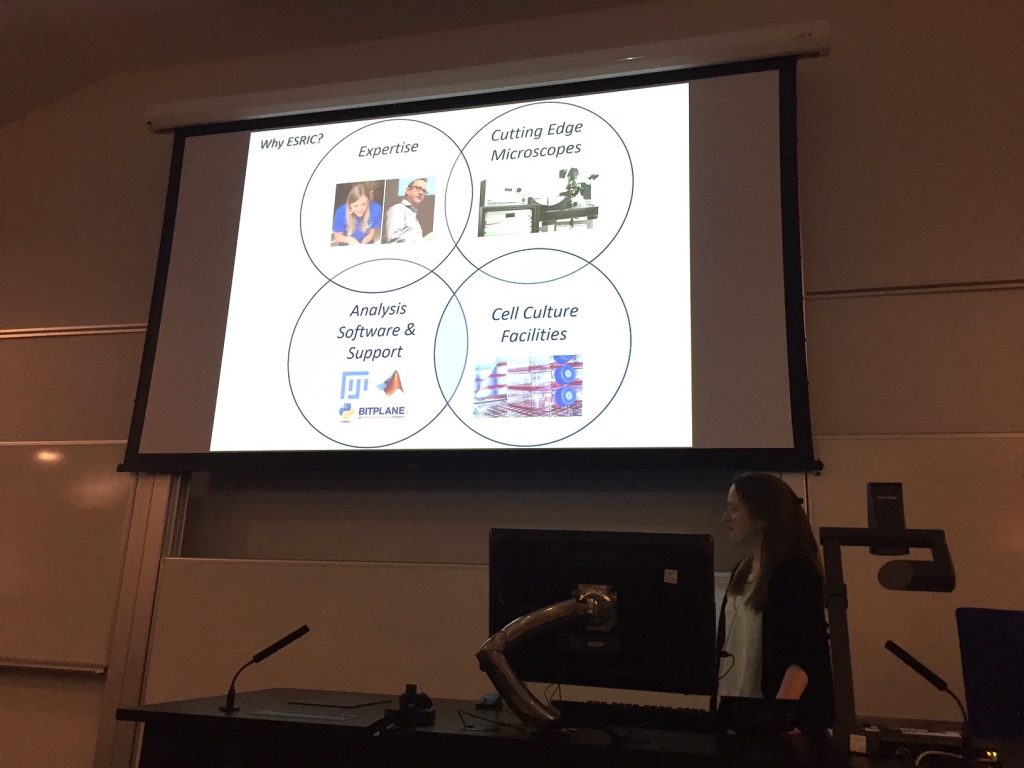
Dr Charlotte Buckley of University of Strathclyde provides an overview of ESRIC facilities at the Super-Resolution Clinic. These include: cutting edge microscopes; analysis software and support; cell culture facilities; and, critically, in-house expertise. Image courtesy of Ali Dun, ESRIC.
Matthew Broadhead of St Andrews University presented an exceptional use-case scenario in which he used the ESRIC super-resolution imaging facility to explore synaptic nanostructure in the mouse brain. This study utilised a protein known as PSD95 that is enriched in the synapse – what Matthew refers to as the most complex organelle in the brain. ESRIC imaging allowed Matthew to explore the distribution of fluorescent PSD95-GFP and this was used to measure the size of synapses, and furthermore to classify synapses as types of nanocluster. A striking observation was that synapses are tripartite and are composed of presynaptic and postsynaptic terminals – as one would expect – but additionally include contact with astrocytes. Astrocytes are star-shaped glial cells in the brain, and this finding is now being explored further in the context of human tissue and iPS cells to determine the role of the astrocyte in synaptic nanostructure and function.
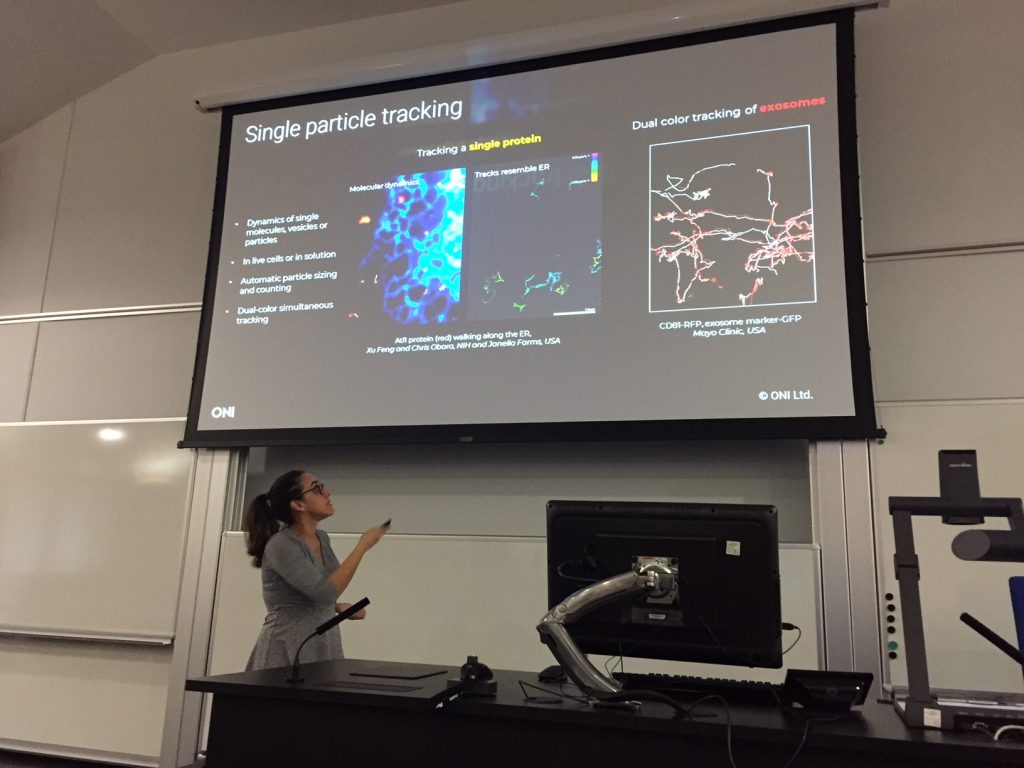
Dr Ana Raquel Periera of Oxford Nanoimaging presents the Nanoimager – a desktop-sized microscope that can generate confocal, SIM, and STORM images. At the ESRIC Super-Resolution Clinic, Ana explained how the Nanoimager can be used to track single proteins and multiple exosomes in living cells. Image courtesy of Ali Dun, ESRIC.
GigaScience has strong interest in publishing datasets with high re-use potential and has published relevant research such as five complete, freely available SIM datasets from human and bovine cells, and mammalian tissue. In addition, GigaScience also has published and hosts four complete, freely available single molecule super-resolution microscopy (SMLM) datasets on YFP-tagged growth factor receptors expressed in a human cell line. These datasets are of great interest to the computational biology and machine-learning community, who wish to develop software tools to analyse nanostructure in cells and tissues. Size limits are not an issue when we can host everything in our GigaDB repository, a recent example showcasing this being the 4TB of immunostaining and simultaneous optical (calcium imaging) and electrical (micro-electrode array) recording we just published. We also have plugins and widgets to host and enable interaction with 3D models (see GigaBlog for more details).
If you have super-resolution microscopy or other large-scale imaging datasets that you wish to submit to GigaScience, please get in touch.
Further Reading
Lukeš T, Pospíšil J, Fliegel K, Lasser T, Hagen GM. Quantitative super-resolution single molecule microscopy dataset of YFP-tagged growth factor receptors. Gigascience. 2018 Mar 1;7(3):1-10. doi: 10.1093/gigascience/giy002.
Pospíšil J, Lukeš T, Bendesky J, Fliegel K, Spendier K, Hagen GM. Imaging tissues and cells beyond the diffraction limit with structured illumination microscopy and Bayesian image reconstruction. Gigascience. 2019 Jan 1;8(1). doi: 10.1093/gigascience/giy126.
Bar El Y, Kanner S, Barzilai A, Hanein Y. Calcium imaging, MEA recordings, and immunostaining images dataset of neuron-astrocyte networks in culture under the effect of norepinephrine. Gigascience. 2019 Feb 1;8(2). doi: 10.1093/gigascience/giy161.