GigaScience at Neuroinformatics 2019

The view from the Marie Sklodowska Curie Museum, where Marie lived as a child.
This year’s Neuroinformatics 2019 meeting took place in the beautiful and historic city of Warszawa (Warsaw). Warszawa remains a pilgrimage city for scientists, with arguably its most famous resident Marie Sklodowska Curie being the first woman to win a Nobel Prize, and the only person ever to win the Nobel Prize twice in two different scientific fields. Cross-disciplinary integration is a major theme in neuroinformatics and this was the perfect setting for the yearly meeting. GigaScience Data Scientist Chris Armit was there and was amazed by the progress that has taken place through the integration of neuroscience and informatics. We’ve attended and supported a number of these International Neuroinformatics Coordinating Facility (INCF) organised meetings in the past (see this write up of the 2015 meeting from Nicole), and this year it was great to see some more general open-science related talks covering community building, preprints, RRIDs, and FAIR data.
Henry Kennedy pointing out many of the essential differences between humans & machines. The disk arrays in his slide are spaced in a way to stop them overheating, whereas us humans do not suffer from this issue.
Connectomics and the Ghost in the Machine
Connectomics (a topic we’ve highlighted in Review articles) was a core theme on Day 1 of the conference. In his outstanding plenary lecture, Henry Kennedy of INSERM succinctly explained the problem with our understanding of cortical connections in primates, which is that the data are collated from multiple sources and in so doing we underestimate the number of connections in the primate brain. Henry compares this to our understanding of the mouse brain, where all cortical areas are known to connect to other cortical areas. Henry’s way of addressing this issue is to use distance-weight relationships to untangle the strength of cortical connections, and this approach generated data that allowed him to state that in the cortex “no area is hugely more connected than others”. However, in Henry’s analysis an important exception to this was the claustrum, which is a thin structure that connects to cortical and subcortical regions. The claustrum shows strong connectivity across all areas of the cortex, and these findings suggest that the claustrum may be the hub that projects to all cortical areas. Henry further suggests that the claustrum may indeed represent a “lost layer” of the insular cortex. I was intrigued by these observations, as they are reminiscent of Francis Crick and Christof Koch’s ideas on the fundamental role of the claustrum in consciousness (Crick & Koch, 2005). Crick & Koch based their hypothesis primarily on the curious anatomy of the claustrum, which appears to connect to all regions of the cortex. Henry’s research would appear to support this early hypothesis and highlight the potential importance of a highly connected claustrum in conscious perception.
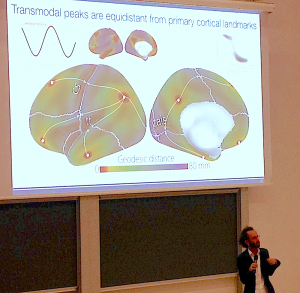
Daniel Margulies explains the importance of geodesic distance in identifying the spatial relationship between transmodal peaks & primary cortical landmarks.
This plenary lecture was a tough act to follow, but Daniel Margulies of CNRS provided invaluable insights into what we can understand using non-invasive approaches on human subjects. Daniel investigates functional connectomics data and explained that, with neuroanatomy alone, it may be difficult to see patterns. However, by using connectivity space – which utilises graph theory – patterns emerge that can then be mapped onto the anatomical model and can provide important insights into cortical connections. I was very taken by Daniel’s approach of exploring graph theory data, which is divorced from neuroanatomy, and re-integrating it with regional, anatomy-based models. Graph theory, whilst very powerful, can sometimes fall into the trap of being over interpreted, especially if dimension reduction was incorporated into the analysis. I came across this issue before in the field of single-cell transcriptomics, with GigaScience Editorial Board Member Wolfgang Huber of EMBL-Heidelberg explaining the potential pitfalls of dimension reduction whereby they “can create one-dimensional (“time-like”) patterns that have little to do with the data-generating process” (see GigaBlog on VIZBI2019). Daniel Margulies offered similar insights and cut the phrase, “it is very tempting to be driven by the algorithms here”. The validity of the graph theory approach is immensely improved by visualising these data in the context of neuroanatomical models, and opens up the possibility of integrating graph theory data into a multimodal spatial framework that is more intelligible to the neuroscience community. In support of this, Daniel highlighted the importance of geodesic distance, rather than Euclidean distance, in discovering patterns in the human brain. To illustrate with one example, it was observed that in geodesic space, transmodal peaks are equidistant from cortical landmarks such as the central sulcus and the callocal sulcus, and this suggests local intrinsic connectivity as an organising mechanism in the human cerebral cortex.
FAIR Principles and Next-Gen Neuroinformaticians
GigaScience Editorial Board Member Carole Goble offered a profound insight into how neuroinformaticians can ensure that their data is Findable, Accessible, Interoperable, and Reusable (FAIR). FAIR is a Digital Object Research Commons and can be thought of as an ecosystem of pooled community resources with many entry points. In such a way, it is distinct from a database / data warehouse that may have only a single entry point. Carole pointed out that in information science, “the tendency to overengineer is strong” and instead highlighted the importance of simple, scalable measures that will be of immense significance in ensuring that data are FAIR. An example that Carole recommends is schema.org that enables structured data descriptors to be included in web pages in a low-barrier universal markup. Of which we at GigaScience are well aware, having integrated it into our GigaDB repository metadata and seen the increase in discoverable as a result.
 I was delighted to meet with Samir Das (pictured) of the Canadian Open Neuroscience Platform (CONP). Samir holds the unique title of being the individual who has received the most brain scans in the world. A volunteer on many brain imaging projects over a number of years, Samir has undergone over 200 MRI scans and was quick to point out that some GE scanners are a lot noisier than others! Samir’s talk focused on dissemination of FAIR datasets and analysis pipelines in the context of CONP, and he highlighted the utility of Boutiques – which was published in GigaScience – and which is a system to “automatically publish, integrate, and execute command-line applications across computational platforms”. Samir’s excellent presentation included bold statements such as: “Data has to be distributed”; and “Tools will need to be versioned”. These represent, to my mind, excellent working practice and invaluable advice for the neuroinformatics community.
I was delighted to meet with Samir Das (pictured) of the Canadian Open Neuroscience Platform (CONP). Samir holds the unique title of being the individual who has received the most brain scans in the world. A volunteer on many brain imaging projects over a number of years, Samir has undergone over 200 MRI scans and was quick to point out that some GE scanners are a lot noisier than others! Samir’s talk focused on dissemination of FAIR datasets and analysis pipelines in the context of CONP, and he highlighted the utility of Boutiques – which was published in GigaScience – and which is a system to “automatically publish, integrate, and execute command-line applications across computational platforms”. Samir’s excellent presentation included bold statements such as: “Data has to be distributed”; and “Tools will need to be versioned”. These represent, to my mind, excellent working practice and invaluable advice for the neuroinformatics community.
The session on Training in Neuroinformatics was a particularly welcome addition to this year’s meeting, and relevant to the Brainhack series we have been publishing and covers the outputs of lot of student projects. William Grisham of UCLA explained that, as an educator on a 10-week neuroscience course, he is unable to accomplish cortical tracing experiments or similar wet lab neuroscience experiments that require longer time scales. However, if his students can gain access to data, then the informatics skills necessary to analyse real datasets can be incorporated into a 10-week programme. This in itself highlights the importance of FAIR principles in neuroinformatics. William has played a crucial role in delivering the UCLA iNeuro web resource, which aims to prepare “a workforce to meet the challenge of large-scale data in neuroscience”. William additionally brought to our attention the INCF Training Space developed by Mathew Birdsall Abrams of the INCF secretariat. This is an incredible open resource that has a vast array of course categories including: Computational Neuroscience; Ethics; Science Management; Neuroinformatics; Data Science; Brain Medicine; Neurobiology; Computer Science; and Tools. Video presentations of exceptional quality are provided on all of these topics with the speaker’s name listed alongside the presentation. Mathew explained to me that as part of the INCF Training Space vision, speakers could be contacted about the content of their online tutorials. In this way there is a feedback mechanism in place to ensure that video material is coherent and intelligible to the target audience. This is an incredible achievement, and is perhaps the best example of online training suited to a cross-disciplinary field that I have come across.
The Dance of the Honeybee
It was great to see another GigaScience Editorial Board Member at Neuroinformatics 2019, Thomas Wachtler of the Ludwig-Maximilians-Universität München, and his colleague Hidetoshi Ikeno of the University of Hyogo, presenting at the meeting. Thomas and Hidetoshi have been collaborating to understand the neurobiology of the famous waggle dance of the honeybee. This is a wonder of the natural world and is the means by which honeybees communicate the direction and distance of flowers with nectar and pollen. I had naively assumed that the waggle dance was a visual display, and so I was quite taken aback when Thomas explained that, “it is dark in the hive”. The waggle dance is not a visual phenomenon, but rather an auditory phenomenon whereby honeybees speak to each other using air vibrations generated by “oscillating their abdomen and beating their wings” (Kumaraswamy et al., 2019). At the conference, Thomas and Hidetoshi presented a pipeline that utilised anatomical and physiological data that were recorded in one lab, and that were shared across labs for collaborative analysis and to ensure the robustness of the pipeline. In particular, I was extremely impressed by the Reg-MaxS-N registration tool that was used to register individual honeybee neurons using morphological criteria. Based on their analysis, the authors report that there are morphological differences in vibration-sensitive neurons of forager honeybees and newly emerged adult honeybees, and that this may highlight a refinement of network processing of air vibration signals. These findings may represent the key to unlocking our understanding of how honeybees ‘listen’ to the waggle dance.
Atlas Informatics – The Importance of Spatial Frameworks
Jan Bjaalie highlighted the multi-scale and multi-level challenges that are being addressed by the Human Brain Project.
Jan Bjaalie (University of Oslo) presented the Human Brain Project, which includes an impressive and versatile web portal that uses knowledge graphs to enable users to undergo the journey, to use Jan’s phrase, from “Data to knowledge”. I was deeply impressed by the integrity of the curation process that Jan outlined. As Jan stated in his talk, the data curation process “is completely concrete”. The HBP website includes data from 405 scientists, and over 90% of submitting authors are themselves members of the Human Brain Project. A key feature of HBP is that section data from submitting authors are spatially warped onto Allen Brain Atlas anatomical models. This enables spatial similarity queries to be performed on spatially mapped data and offers a method of exploring, for example, coexpression in the human brain. As Jan pointed out, in this spatial framework the research data is not warped, and is instead left intact. Rather, it is the atlas images that are warped and these provide the necessary anatomical context for understanding the raw data. As GigaScience are purists of CC0 public domain waivers for data, I did notice that some of the Creative Commons Licenses that were ascribed to submitter data were CC-BY-NC (non-commercial) and CC-BY-ND (no derivatives) and I did ask Jan whether this will impact on reuse of the original data. Jan made the case that for the HBP resource, the emphasis is on providing details of the license chosen by the submitting authors rather than enforcing a license that is more compatible with reuse.
Alexander Woodward of RIKEN CBS presented a 3D marmoset atlas as a common space, and additionally a population average based on 25 scans. The average atlas utilised Advanced Normalisation Tools (ANTS). ANTS was orignally developed by Brian Avants, Jim Gee and colleagues at the University of Pennsylvania and remains one of the most impressive automated methods for spatially mapping high-contrast brain imaging data generated by CT and MRI. I was aware that it was possible to add fiducial points to these datasets to ‘anchor’ and help align brain images prior to the ANTS-based elastic (diffeomorphic) transformation. However, from discussions with Alexander Woodward and his colleague Piotr Majka (Nencki Institute, Warszawa) I became aware of a more recent modification to the ANTS protocol whereby Jim Gee, working with Piotr Majka’s group, has now enabled anatomical domains – such as brain ventricles – to be used as ‘anchors’ and this greatly enhances ANTS-based registration around the anchored regions. This is an incredibly important refinement as more accurate spatial mapping will increase the granularity of spatially registered datasets.
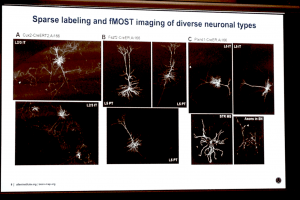
Michael Hawrylycz of the Allen Institute for Brain Science showcased fMOST (fluorescence micro-optical sectioning tomography) imaging in tissue-cleared transgenic mouse brains, which allows individual neurons to be imaged in situ at cellular resolution.
The last speaker of Neuroinformatics 2019 was Michael Hawrylycz of the Allen Institute for Brain Science who gave a visually stunning overview of the history of the Allen Brain Atlas from its inception as a mouse brain atlas of microanatomical level in situ hybridisation data towards its move to single cell-resolution data. What I thought was most exciting here was the tantalising glimpse of fMOST (fluorescence micro-optical sectioning tomography) imaging in tissue-cleared mouse brains as a means of directly viewing brain-wide connectomics at the cellular level. In one of the example images, the claustrum was labelled and this highlights a technological advancement that may offer profound insights that build upon the aforementioned observations made by Henry Kennedy.
A further profound insight offered by Michael was that there will be a need for Deep Learning to recognise the myriad cells and cell types in these incredibly information-dense images.
The Developing Brain – a Future Focus for INCF?
Although there was not a dedicated session on the developing brain, this was a theme that kept appearing in panel discussions. Questions from the audience related to the role of developmental processes in creating the architectonic relationships that we see in the adult brain, and there was further discussion on the importance of gradients of gene expression in creating architectonic boundaries. In addition, I noted that there were questions relating to the genesis of the first generation connectomic hubs that appear in embryonic development. In this respect, I found the words of Claus Hildetag (Universitätsklinikum Hamburg-Eppendorf) – that connectomic hubs may simply represent those neurons that are “first to the party” – incredibly insightful, and a challenge to the developmental neurobiology community. At future INCF meetings, I hope we can see more of a focus on the biology and modelling of developmental mechanisms in the brain so that we can find out whether this hypothesis holds up to deeper scrutiny.
Neuroinformatics 2019 was organised by the INCF who have a permanent secretariat based in the Karolinska Institute, Stockholm and who promote the implementation of neuroinformatics, and promote data reuse and reproducibility in brain research.
#NI2019 was a success! We at INCF are very grateful to all the speakers, local organizers, sponsors, and attendees for making it a truly #neuroinformagical event! pic.twitter.com/Ycmzn0rafq
— INCF (@INCForg) September 5, 2019
Neuroinformatics 2020 will be in Seattle on August 17-21 and will be hosted by the Allen Institute for Brain Science.