What’s your poison? Venom composition of 26 deadly snakes
The Center for Antibody Technologies headed by Professor Andreas Laustsen-Kiel (Technical University of Denmark) used high-throughput methods to systematically analyze the venoms of the 26 most deadly snakes in sub-Saharan Africa. The results are now published in Gigascience.
Each year, around 500,000 people in sub-Saharan Africa suffer from snake bites, causing an estimated 7,000 to 20,000 deaths. Many snake species native to the region, such as the feared black mamba, are classified as species of the highest medical importance by the World Health Organization. Using systematic approaches and state of-the-art methods to better understand the composition and function of these snakes’ venoms is a medical priority (see also this recent Review Article in Gigascience, focusing on the role of genomics in this effort, and this article for a general perspective on modern venomics).
Proteomics of 26 deadly African snakes
The snakes investigated in the new proteomic and functional study belong to two families, elapids – including, among others, the black and green mambas and the ring-necked spitting cobra, also called the rinkhals – and vipers such as the Puff adder and the Gaboon viper.

The composition and function of snake venoms is complex and varies a lot from species to species.
The authors describe a general pattern with elapid venoms containing large amounts of a class of proteins called “three finger toxins”, which act by blocking neuronal transmission or by killing cells; as well as phospholipases A2 (PLA2s), a class of enzymes that is found in many animal venoms. The viper venoms, on the other hand, are dominated by a different protein mix, including PLA2s, but also substantial quantities of other enzymes such as Snake Venom Metalloproteinases and Snake Venom Serine Proteinases.
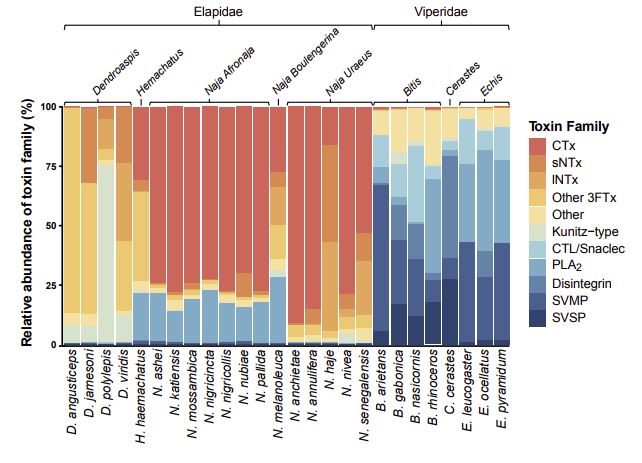
Venom compositions of most of these snakes have been described before, but the venoms of two species – of Anchieta’s cobra (Naja anchietae) and of the white-bellied carpet viper (Echis leucogaster) – are characterized for the first time in the new GigaScience study.
An integrate approach to understand venom composition and function
The major advance of the work, however, is the parallel processing of samples from 26 snakes in the same high-throughput pipeline; combined with a range of experimental approaches to functionally characterize many venoms in parallel, in a standardized setting. In contrast, previous studies on the venom compositions of snakes from sub-Sahara Africa have typically been performed in separate studies with only one or a handful of species each, and often with little or no data on functional aspects. The previous studies also used variable protocols, making it difficult to reconcile and compare data from different origins.
The new integrated approach demonstrated in the Gigascience article, including 26 snake species and a range of functional assays, provides a solid foundation for further studies of snake biology and the development of new antivenoms.
Read the GigaScience article:
Nguyen GTT; O’Brien C; Wouters Y; Seneci L; Gallissa-Calzado A; Campos-Pinto I; Ahmadi S; Laustsen AH; Ljungars A High-throughput proteomics and in vitro functional characterization of the 26 medically most important elapids and vipers from sub-Saharan Africa. GigaScience 2022. https://doi.org/10.1093/gigascience/giac121
Supporting data in GigaDB: http://dx.doi.org/10.5524/102333
Read related posts on GigaBlog: