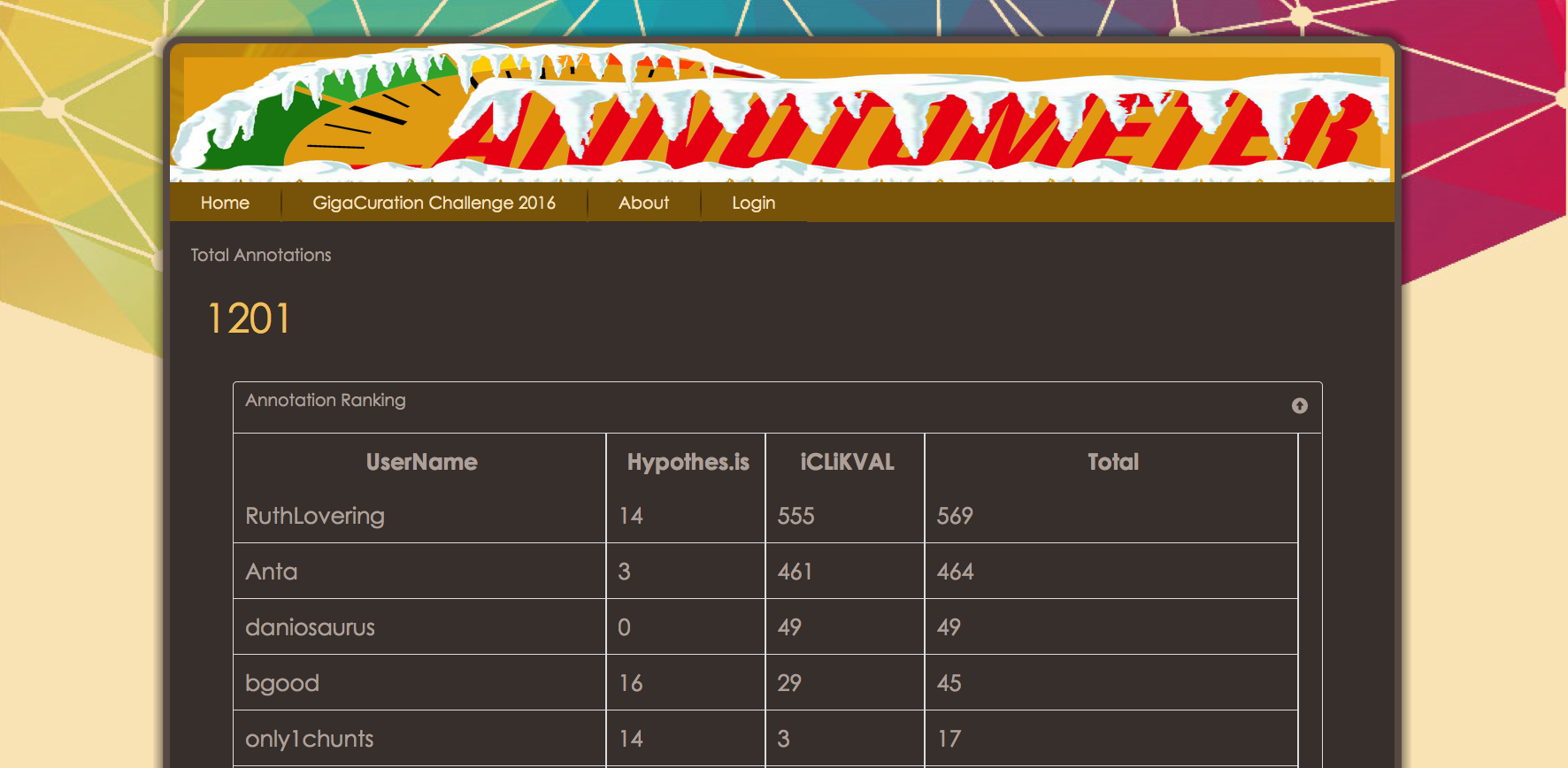
The Annotometer Results Are In…
The Leader of the Curation Pack.
At GigaScience we are keen to maximise the use and utility of our published data, narrative and other research objects. Curation is key to doing this, and on top of the role of professional biocurators, the “many-eyes” approach of harnessing crowdsourced community annotation has been much discussed, but not realised other than a few notable exceptions leveraging wikipedia (wikidata, wikigenes, etc.). At this years Biocuration Society (Biocuration 2016) meeting we decided to role up our sleeves and try to seed annotation of our and others data resources through a “GigaCuration Challenge”. You can see more in our previous GigaBlog posting, but the carrot provided was the prize of an iPad for the person making the greatest number of annotations at the meeting. Being part of the “Annotating All Knowledge” consortium and implementing Hypothes.is annotation plugin on our GigaDB repository that was one obvious too to use here (see announcement blog and Q&A with our new Ed Board member Maryann Martone here). We also encouraged and counted annotations from iCLiKVAL, a web-based tool that accumulates annotation information for all scientific literature and other media such as YouTube, Flickr, and SoundCloud.
The new tool that we developed especially for this challenge, called the Annotometer, worked well for its first real test, and we have gathered some valuable feedback and experience to fuel its improvement for future competitions. The live leader-board worked consistently from start to end, and froze when we ended the competition, the final table can still be found on the annotometer.com website. The competitive nature of a live leaderboard has driven other science competitions such as the Sequence Squeeze Challenge (see the commentary in GigaScience from the organisers for lessons learned from that), and is well practised in open innovation challenges, making the kaggle datascience competitions so popular for example.
If you build it, will they come?
We learned several lessons from this event;
1- A five minute introduction was insufficient to really get things started, next time we will try to get a longer time slot from the organisers.
2- Professional BioCurators are a surprisingly shy bunch, with only 22 out of ~200 attendees registering for the challenge.
3- Four days was perhaps too long for a single challenge, so in future we may consider running daily challenges with prizes for each day.
4- Feedback from non-participants suggested that by not specifying exactly which articles to curate (i.e. leaving to user choice) was what put them off!
5- The number of curations done on any of “Getting-started” suggested articles, was approximately, zero, indicating that none of the users bothered to look around the Annotometer website!
Congratulations to Ruth Lovering from UCL (pictured when collecting the prize Chris), who after a close fight at the top, carried out 569 annotations during the meeting.  The results on the leaderboard may look like there was a massive bias to iCLiKVAL over Hypothes.is (1197 vs 88 annotations), however we think there are multiple factors that can account for that difference in usage. Firstly, as the audience were all curators, these are people that are familiar and comfortable with structured data, iCLiKVAL provides a tool for adding triplet annotations, the exact sort of thing that curators like. Secondly, the hypothes.is application is not yet available on any browser other than Chrome, and is not yet fully functional on mobile devices. Both these issues were raised as feedback and are indeed areas that hypothes.is are working on now. Finally, iCLiKVAL is designed to be quick and easy to add short annotations, compared to hypothes.is which is designed to allow conversations over any webpage. For this reason, it maybe necessary for us to implement a new scoring system for future challenges (watch this space).
The results on the leaderboard may look like there was a massive bias to iCLiKVAL over Hypothes.is (1197 vs 88 annotations), however we think there are multiple factors that can account for that difference in usage. Firstly, as the audience were all curators, these are people that are familiar and comfortable with structured data, iCLiKVAL provides a tool for adding triplet annotations, the exact sort of thing that curators like. Secondly, the hypothes.is application is not yet available on any browser other than Chrome, and is not yet fully functional on mobile devices. Both these issues were raised as feedback and are indeed areas that hypothes.is are working on now. Finally, iCLiKVAL is designed to be quick and easy to add short annotations, compared to hypothes.is which is designed to allow conversations over any webpage. For this reason, it maybe necessary for us to implement a new scoring system for future challenges (watch this space).
As regulars at Biocuration (see writeups from 2012 and 2015), it was great to be a formal sponsors for the first time. We are in the process of tiding up the annotometer code on github, and will write things up/describe it better to encourage its re-use. We also gathered (generally positive) feedback at the “Annotating All Knowledge” working group at the FORCE2016 meeting in Portland, where Nicole presented hot-off-the-annotometer results in the user cases track (slides here).
