2023 in review: Gigabyte journal coming of age, and more
It’s December, the festive season and the end of year are approaching fast – and it’s time for our traditional look back on the past 12 months at GigaScience Press. Once more, we are pleased with the view in the rear mirror. In its 11th year, GigaScience again published exceptional “big data” science (read on for examples). And GigaScience’s younger sister GigaByte was taking significant steps forward – you could say we witnessed its coming of age this year.
To start with an important milestone reached in 2023, GigaByte is now indexed in PubMed, one of the major searchable databases of biomedical citations. In addition, the journal is now indexed by PMC (PubMed Central), a free digital archive of the full-text of Open Access articles.
And in really recent news, we have also heard from another literature database, Scopus. Watch this space, details will be announced soon!
Being indexed in these databases is an important boost for the visibility and findability of GigaByte’s papers. In addition, the automatic inclusion into PMC is also necessary for many researchers to meet their research funders’ policies and mandates on Open Access. Indexing a journal requires a solid and high-quality management of metadata, which we put in place together with our partner RiverValley (read the blog for details).
Publish, Review, Curate
Over the last year we also improved the GigaByte experience in other ways: In addition to innovative features such as multilingual articles, GigaByte is now also offering a highlighted editor’s assessment of each reviewed preprint. The editor’s endorsement helps readers to understand the importance of the work, while also giving insights on the review process. This new feature is part of the ‘Publish, Review, Curate’ (PRC) peer-review model, using eLife’s Sciety platform. Via Sciety we also share our open, signed peer reviews for preprints that are published in GigaByte and GigaScience.

GigaBlog going rogue
Talking about indexing, also this very blog, GigaBlog, improved its findability and long-term archiving. GigaBlog is now archived via Rogue Scholar, a platform operating under the motto “science blogging on steroids”. In practical terms this means that Rogue Scholar provides full-text search, long-term archiving, DOIs and metadata for Open-Access science blogs such as ours. In the 12 years of its existence, GigaBlog has published numerous author Q&As, accessible summaries of research highlights, conference reports, and guest articles, touching on scientific and publishing-related topics. More than 300 posts in total.
With the help of Rogue Scholar, this research-related (but not peer reviewed) content is now becoming part of a wider network of archived and curated science blogs. With the ongoing troubles at corporate-hosted social media platforms such as ex-Twitter, it is reassuring to have our own self-hosted space for discussion and outreach. By the way, talking about social media, you can now also connect with us via Mastodon and Bluesky.
Vectors of Disease

But back to the peer reviewed content in our journals. In 2021, GBIF (the Global Biodiversity Information Facility) and GigaByte worked together to launch a new thematic series: Sponsored by a special program of the World Health Organization, we published a series of papers describing datasets on vectors of human diseases. This thematic series mobilized more than 500,000 occurrence records and 675,000 sampling events from more than 50 countries. Building on this success, we continued the fruitful partnership with GBIF for a second call for papers in 2023 – now including more countries (from Asia and Africa) and also new taxa, including snails and rodents.
The second part of the thematic series on disease vectors also includes a real gem for natural history nerds: A paper on the field notes of Dutch entomologist Johanna Bonne-Wepster (1892-1978), which now have been digitized, and are therefore made available for 21st century public health work (Read the blog post here).
Bibiche Berkholst from the Naturalis Biodiversity Center commented on the collection:
“Bonne-Wepster collected and documented a huge amount of mosquitoes in her lifetime. We were able to digitise over 10,000 of those, which took us many months. Each mosquito had to be checked against Bonne-Wepster’s field books. It feels very satisfying that this important collection has been digitized, and I hope that the remarkable history of Bonne-Wepster will inspire many female amateur entomologists like myself.”
Don’t miss this video to see the actual mosquitoes and the curators at Naturalis who digitized the collection:
Gigasnakes

Another thematic series in GigaByte also deals with animals that often are a public health concern – Snakes! Genomic data helps to understand the evolution of snake venoms and also to study the evolution and diversification of these fascinating reptiles.The series in GigaByte presents genomes and annotations of snakes such as the Chinese cobra, the brown spotted pit viper and others.
With all the innovations and exciting developments at our young journal GigaByte, we didn’t neglect its older sister GigaScience, which had another strong year.
To highlight just a few of the 2023 publications:
Meet Cooinda the dingo
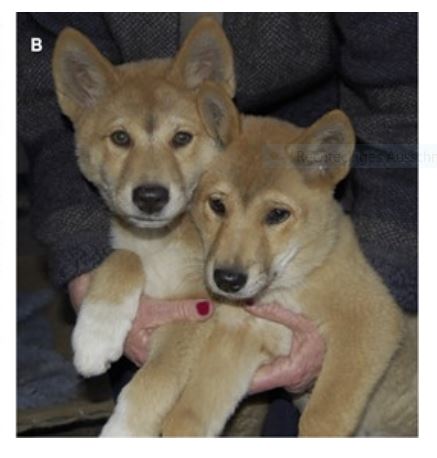
In March, an Australian team led by Bill Ballard (University of Melbourne) published rich multi-dimensional data for one Alpine dingo named Cooinda. High-quality genome sequences, molecular biology, magnetic resonance imaging and morphometric techniques were all brought together. Cooinda can now serve as a reference point for detailed evolutionary comparisons of Dingoes at all levels – including morphology, genetics and molecular biology (Read the paper here, and the blog post here).
Metabolite profiles for 1600 plants
We are especially happy if we can help scientists to open up datasets that previously were unavailable for the wider scientific community. A very nice example for this mission was a paper by Pierre-Marie Allard and colleagues, presenting a large library of plant metabolites.

Pierre Fabre Laboratories, a French pharmaceutical and cosmetics company, constituted a collection of botanical samples over two decades. This collection is one of the largest private plant libraries in the world. It contains over 17,000 unique samples, including some rare species and covering a diverse range of botanical families. The GigaScience paper starts to open this resource up by sharing metabolite profiles for 1600 plants from the collection – data that can also be useful for drug discovery, for example (Read more on GigaBlog, here)
Amazing Amazonian Butterflies
We are more and more selective regarding the types of genome sequencing work that we consider for publication in GigaScience, given the technical progress in the field. Genomic work that is more incremental and not coming together as a “complete story” is better suited for the sister journal GigaByte. However, we are sympathetic towards authors who provide high-quality reference genomes for taxonomic groups that have been overlooked so far by the genomics community.

A prime example for this type of paper is the article on three amazonian butterfly species: Morpho butterflies are emblematic species of the Amazonian rainforest, loved for their metallic shades of blue and green. Despite their conspicuous looks and fascinating biology, large-scale genome sequencing efforts have somewhat neglected them – until Héloïse Bastide and coworkers published not one, but three genomes of the genus in GigaScience (read the article here, and the blog post here, ).
The 300$ HARU device

Also in the area of methods, we are especially keen to publish work that fosters open science practices and helps scientists with few financial resources to contribute to life science research, without having to rely on expensive, proprietary soft- and hardware.
One nice example of how this mission can be achieved is a paper by Hasindu Gamaarachchi, presenting their “HARU” system for selective sequencing, to be used alongside the MinION handheld sequencer. As Dr. Gamaarachchi explained in our author Q&A, their tiny $300 device is “two times faster than a 30,000 $ 36-core server, at a fraction of power consumption”.
Birthday party and other travel business

As always (well, not counting times of worldwide pandemics) our editors and data curators traveled to conferences and workshops, to keep up-to-date on current progress and to meet our authors, reviewers and editorial board members – and to join our traditional GigaScience birthday party at ISMB, hosted this year in Lyon (France) . Our Editor-in Chief Scott Edmunds noted after coming back:
“This year proved that conferences are properly back, with the highest turnout for the meeting to date: 2100 in-person attendees and 400 watching online. It was great to see lots of familiar faces that hadn’t been back for many years!”
Our birthday celebrations were equally larger than expected, with more than 100 attendees at the end of the night. Also Giga-Panda-macarons proved to be a great hit!
At the International Conference on Genomics (ICG), our Editor-in-Chief talked about Mobilization of Public Health Data – you can watch the talk here:
In addition we were present at lots of other conferences, including PAG (Plant and Animal Genomes) in San Diego, and celebrating 10 years of the Research Data Alliance in Gothenburg, Sweden.
Looking ahead
What will 2024 bring? More scientific progress, that’s for sure – for example, we are going to launch a new thematic series on “telomere-to-telomere” genomes, that is, gap-free assemblies. Until recently, such a genomic tour de force was only attempted for the major model organisms and the human genome. Today, this quality standard is achievable also for less prominent organisms, opening the door for new research that requires truly complete, gap-free genome data.

In any case, science and publishing is not standing still, and with all the progress there are also new challenges ahead, especially seeing how machine learning and AI-based tools change scientific practice.
Submissions involving machine learning methods are increasing in frequency – and they are often a challenge for journals such as GigaScience and GigaByte, which insist that research needs to be reproducible, with transparent methods; and that a submission is only complete with all associated data and code openly available. The “DOME-ML” recommendations ( for Data, Optimization, Model, Evaluation) are one effort to ensure that submissions using supervised machine learning methods are following best practice. In addition to referring our authors to these guidelines, we also started a trial where authors are asked to complete a set of standardized questions on their methods and data, provided by the DOME initiative. As part of the trial, we host these “DOME annotations” in our database GigaDB, increasing transparency for our readers and reviewers.
Another trial we initiated this year is the use of Data-SEER, an AI tool that provides support for data-related aspects of new submissions. The Data-SEER report helps authors and editors to spot areas for improvement – for example to make sure that standard data types come with appropriate accession numbers.
New technologies come with risks and opportunities, and this is also true in the world of scientific publishing. In 2024, we will keep an eye on the risks, but continue to look at opportunities to make science ever more open.
We thank our readers and authors for their continued interest in GigaScience and GigaByte, and our reviewers and editorial board members for their support!
Merry Christmas , happy holidays, and happy new year!

References:
Pasquale Ciliberti, Astrid Roquas, Becky Desjardins, Bibiche Berkholst, Frank Loggen, et al. Digitizing the Culicidae collection of Naturalis Biodiversity Center, with a special focus on the former Bonne-Wepster subcollection, Gigabyte,2023 https://doi.org/10.46471/gigabyte.85
J William O Ballard, Matt A Field, Richard J Edwards, Laura A B Wilson, Loukas G Koungoulos, Benjamin D Rosen et al. The Australasian dingo archetype: de novo chromosome-length genome assembly, DNA methylome, and cranial morphology, GigaScience 2023, giad018, https://doi.org/10.1093/gigascience/giad018
Pierre-Marie Allard, Arnaud Gaudry, Luis-Manuel Quirós-Guerrero, Adriano Rutz, Miwa Dounoue-Kubo et al. Open and reusable annotated mass spectrometry dataset of a chemodiverse collection of 1,600 plant extracts, GigaScience 2023, giac124, https://doi.org/10.1093/gigascience/giac124
Héloïse Bastide, Manuela López-Villavicencio, David Ogereau, Joanna Lledo, Anne-Marie Dutrillaux, Vincent Debat, Violaine Llaurens, Genome assembly of 3 Amazonian Morpho butterfly species reveals Z-chromosome rearrangements between closely related species living in sympatry, GigaScience 2023, giad033, https://doi.org/10.1093/gigascience/giad033
Po Jui Shih, Hassaan Saadat, Sri Parameswaran, Hasindu Gamaarachchi, Efficient real-time selective genome sequencing on resource-constrained devices, GigaScience 2023, giad046, https://doi.org/10.1093/gigascience/giad046