Cone snail genome sheds light on venom evolution
This week in GigaScience we published the genome of the mediterranean cone snail, Lautoconus ventricosus. Cone snails produce a wide variety of powerful toxins and the new chromosome-scale genome assembly opens the door for detailed investigations of their diversity and evolution.
Cone snails are beloved by collectors for their often beautiful shells, but they are infamous because of their powerful venom. As predatory species, they hunt marine worms or even small fish by firing a toxin-charged harpoon (a modified tooth) at their victims. This might also explain why many of the conotoxins are so powerful: the snails need to paralyze their victims quickly, or otherwise their lunch might get away.
Worldwide, there are more than 900 species of cone snails and the venom of certain larger species can kill a human. However, for their sequencing project, Rafael Zardoya and colleagues chose a smaller species, which is harmless to humans: the mediterranean cone snail Lautoconus ventricosus.
“ The species Lautoconus ventricosus is the cone snail endemic to the Mediterranean Sea and the Atlantic Ocean region near to the Gibraltar strait.”, Zardoya explains, adding: “Although its olive-colored shell is not as stunning as those of cone snails from the Indo-Pacific region, it is one of the earliest cone snails described and has a long taxonomic history.”

Manuel Tenorio, also an author of the paper, explains the background to the project: “We have been working on different aspects of cone snail biology for more than a decade. We have focused our interests on the West African region, a hotspot of cone snail diversity, but also work on cone snails from other parts of the world. Sampling the local species, abundant in southern Spain and Portugal was straightforward. Furthermore, we managed to keep specimens of L. ventricosus alive in aquarium where they have adapted pretty well. We feed them with live bait, and they have been able to reproduce in captivity. This will certainly be very useful for future developmental biology studies, currently in progress.”
Conotoxins (that is, the venoms of cone snails) are a family of usually smallish peptides of around 10-30 amino acids in length. They are very precise in their action and can be sorted into groups according to their molecular targets, various ion channels. Their precise and potent effects make conotoxins also interesting for pharmacological researchers. For example, one conotoxin-derived drug (Ziconotide) is approved for medical use and is given to patients who need powerful pain release. Ziconotide blocks one specific type of ion channel, the N-type voltage gated calcium channel, and thereby inhibits the release of neurochemicals in the nervous system. Used as a pain reliever, the substance is magnitudes more powerful than morphine.
What makes conotoxins so fascinating is their diversity: not only is there a large variety between species, but also the venom gland of one individual specimen typically contains not one but dozens, if not hundreds, of structurally different peptides, with a large variety of precursors encoded in the genome. Why is that so? Tenorio explains:
“Cone snails belong to the superfamily Conoidea, a remarkably biodiverse group of venomous marine invertebrates comprising more than 10000 species. Conoidea are successful top predators in the marine realm thanks to the venom, a key evolutionary innovation. They have diversified through recurrent evolutionary radiations. Venom peptides diverge at a very high rate in a continuous arms race (red queen strategy) to adapt to changes in their targets in the prey. “
Seen through the lens of evolutionary theory, whenever there are predators and prey, there’s usually an arms race. Producing a cocktail of different, fast-changing toxins gives the predators a temporary advantage. A trajectory towards resistance against one stable toxin might exist, but evolving lasting countermeasures to a diverse, rapidly changing spectrum of toxins is not so straightforward.
To add further complexity, functional diversity arises also due to downstream modifications at the transcriptional and protein level. Therefore, to understand the function and evolution of conotoxins, the entire toolkit of modern biology is needed: genomics, transcriptomics, and proteomics. A chromosome scale reference genome, such as the one published now in GigaScience, is a crucial element to bring it all together.
The new genome assembly is one of the largest gastropod genomes sequenced so far, with 3.59 billion base pairs organized in 35 chromosomes. The authors annotated 32,675 protein coding genes.
The Spanish scientists also explored the transcriptome, specifically of the venom gland, and identified 289 transcripts that could be assigned to known conotoxin precursor families. Mapping these transcripts to the reference genome, the overall picture is that venom precursor genes are not clustered, but distributed over the entire genome, although with some traces of past “tandem“ duplication events.
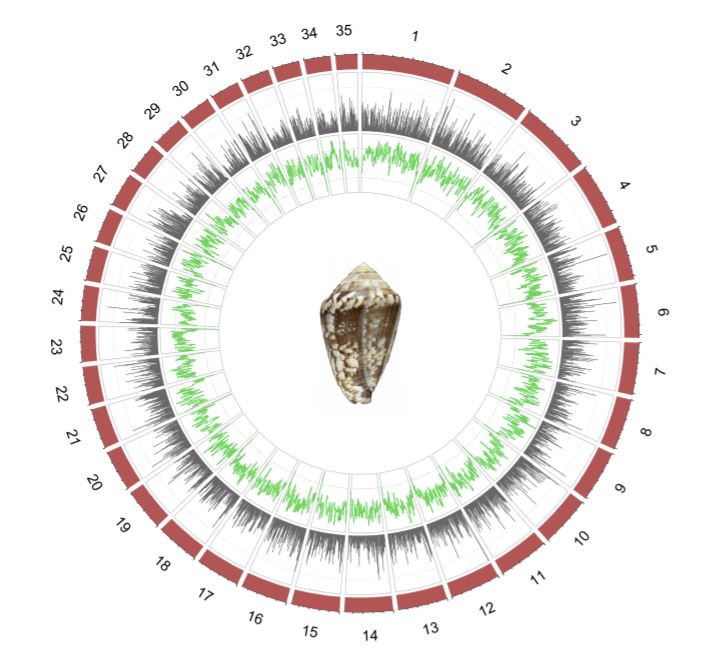
Zardoya: “For many years, it was puzzling how the great diversity of toxins was generated. The genome answers this question. Many gene copies distributed throughout the different chromosomes are producing this diversity, which is later enhanced by post-translational modifications of the peptides. “
Another group recently also published a chromosome-level genome of a different cone snail species, Conus betulinus. Together with the genomes of other venomous animals, such as the Solenodon and the Cobra, evolutionary biologists can now have a closer look at the origins of venoms in various animal branches.
Tenorio: “As more high-quality genomes of venomous species in the different branches of the Animal Tree of Life are assembled, we will be able to discern which venoms are inherited through common ancestry and which ones appear due to convergence in the different lineages.”
References
The genome of the venomous snail Lautoconus ventricosus sheds light on the origin of conotoxin diversity José Ramón Pardos-Blas, Iker Irisarri, Samuel Abalde, Carlos M L Afonso, Manuel J Tenorio, Rafael Zardoya GigaScience, 2021 https://doi.org/10.1093/gigascience/giab037
Data in GigaDB: http://dx.doi.org/10.5524/100892