A Decade of FAIR – and what next? Q&A on FAIR workflows with the Netherlands X-omics Initiative
Marking the 10th anniversary of the formulation of the FAIR principles, we have one of our GigaBlog Q&A’s with Peter-Bram ‘t Hoen, Alain van Gool, Anna Niehues and Casper de Visser from the Netherlands X-omics Initiative and Radboud University Medical Center in Nijmegen, authors of a new paper on publishing FAIR workflows. Giving us their insight into how the FAIR principles for research data can now be applied to research software and computational workflows. And also giving first hand insight into the use of technical platforms helping enable this: RO-Crate and wokflowhub.eu.

The “Jointly designing a data FAIRPORT” Lorentz workshop that formulated the FAIR principles in 2014, see https://www.lorentzcenter.nl/jointly-designing-a-data-fairport.html
The FAIR Principles at 10
This month marks the 10th anniversary of the FAIR (Findable, Accessible, Interoperable and Reusable) principles for scientific data management and stewardship. These were formulated after a Lorentz Center workshop in January 2014 where a diverse group of stakeholders, sharing an interest in scientific data publication and reuse, met in Leiden to discuss the features required in our now more data-driven era. The first-draft of the FAIR Principles was then published on the FORCE11 website for evaluation and comment by the wider community (a process that lasted two years) before the write up was published in the very influential “The FAIR Guiding Principles for scientific data management and stewardship” paper (of which our Editor in Chief was one of the authors). To mark this decade of FAIR, later this month there will be a new Lorentz Center Workshop on “The Road to FAIR and Equitable Science”, alongside a number of conferences and workshops in Leiden and beyond celebrating this milestone, and trying to take the field forward for the next decade.
Coming exactly 10-years from this event it is nicely appropriate having a new paper out in GigaScience applying the FAIR principles to a complicated multi-omics analysis workflow combining data, metadata, and software into a complicated research workflow. This “FAIR Digital Object” providing an important case study and lesson in reproducibility. As a journal focussed on open and reproducible science, we have worked on and published many these sorts of case studies (see our PLOS One paper for a good example), and are keen to integrate these types of best practices into our publishing workflows, and also promote them wider to the research community to aid their wider adoption. The new paper showcases on the outputs of the Netherlands X-omics Initiative investigating shared patterns between multi-omics data and childhood externalizing behavior. Demonstrating the development of a FAIR Digital Object comprising a computational workflow that analyzes and integrates multi-omics and phenotype data and is associated with rich human and machine-readable metadata.
Here authors Peter-Bram ‘t Hoen, Alain van Gool, Anna Niehues and Casper de Visser all from Radboud University Medical Center in Nijmegen tell us in a Q&A the insights learned from this project to hopefully through better light on this next phase of FAIR. Also speaking on behalf of other X-omics initiative members from different institutes in the Netherlands who are co-authoring the paper.
Why is reproducibility important to you, and what lead you to carry out the reproducibility case study here?
As the size of -omics data volumes, and the complexity of the corresponding analysis workflows are increasing, reproducibility becomes more important. Researchers need to be able to replicate each other’s studies and to validate each other’s results. This is particularly important when it comes to the introduction of omics technologies in a clinical diagnostics setting, where health decisions are at stake. Within the context of the Netherlands X-omics Initiative (https://x-omics.nl/), we are building a research infrastructure for reproducible -omics data analysis. With our demonstrator projects we aim to showcase how the X-omics research infrastructure can make -omics research more reproducible. Here, we present one of these finished demonstrator projects, in which we developed a fully reproducible workflow to analyze a complex dataset that combines multi-omics and behavioral data.
People may know about the FAIR principles for research data, but how do they relate to software and computational workflows, and what is a FAIR Digital Object?
The FAIR principles were indeed originally designed for (research) data, but have later on been extended to software with the FAIR4RS principles. These principles can guide anyone who develops research software (including computational workflows) to improve reproducibility and reusability of their work. This is beneficial for multiple parties involved: your scientific tool that you worked hard on for a long time can get more impact, and also others can more easily re-use and combine existing software, saving significant time and effort. FAIR Digital Objects can be any digital resource that adheres to the FAIR principles. Hence, our multi-omics analysis workflow can be referred to as FAIR Digital Object.
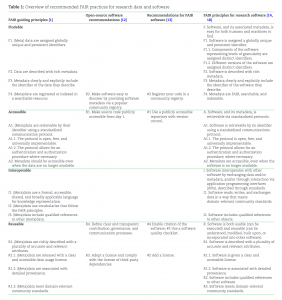
Overview of recommended FAIR practices for research data and software taken from the paper.
Your computational workflow is packaged as an Research Object Crate (RO-Crate), so can you tell us a little about that and what the advantages are in this system for sharing and describing data, datasets and workflows?
The RO-Crate specification facilitates to package your workflow/data/software along with information describing them (metadata), which is crucial to adhere to the FAIR4RS principles. We were able to generate the RO-Crate with a Python library and accompanying documentation. Using the extensive documentation of the RO-Crate, we were able to make our metadata adhere to the Bioschemas specification of workflows as well. Moreover, the RO-Crate can easily be uploaded to WorkflowHub (see https://workflowhub.eu/), where you can register your workflow to make it findable and reusable by others.
Your workflow is also shared via workflowhub.eu (of which GigaScience now endorses to its authors publishing workflows), so what are the advantages of that platform and how did you find using it?
As mentioned above, WorkflowHub is a platform on which you can register your scientific analysis workflows. It supports generation of Digital Object Identifiers (DOIs) which are unique and persistent identifiers for your workflows, and also for every new version. Both workflow versioning and unique identifiers are of key importance for reproducibility. Once we generated our Workflow RO-Crate, it was very easy to publish it on WorkflowHub. However, to update files of the workflow, some manual work was required. This might be avoided by more seamless integration with for example GitHub, but we did not find a way to integrate RO-Crate, WorkflowHub and GitHub in an automated manner. We noticed that there is active development happening for both RO-Crate and WorfklowHub, so we hope this can be possible in the near future. More guidance on making workflows FAIR can be found in our recent paper: “10 quick tips for building FAIR workflows“.
The workflowhub.eu website, now endorsed by GigaScience to archive the computational workflows we publish.What do you hope readers will learn from this case study, and what do you hope they will do with the reproducible examples you produced?
We mainly hope that readers will learn what it takes to makes your analysis workflow completely reproducible, and that there are already many tools out there that you can use for that. Moreover, we experienced that FAIR workflow development is not a task that should be underestimated. However, examples like ours can be used as a template and facilitate the process. Given the importance of reproducible science, it is definitely worth the effort.
You say many of these resources you used for the first time in this case study, so has working in this manner changed how you will carry out your research in the future?
Yes, we have learned a lot during this project. In current and future projects, we can more easily make our workflows FAIR, already during development. Several analysis modules in this workflow are adopted in other projects already.
References:
Niehues A, de Visser C, Hagenbeek FA, et al. (2024) A multi-omics data analysis workflow packaged as a FAIR Digital Object, GigaScience, 13, giad115, https://doi.org/10.1093/gigascience/giad115
Wilkinson, M., Dumontier, M., Aalbersberg, I. et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018 https://doi.org/10.1038/sdata.2016.18
Chue Hong NP, Katz DS, Barker M et al. (2022). FAIR Principles for Research Software (FAIR4RS Principles) (1.0). Zenodo. https://doi.org/10.15497/RDA00068
de Visser C, Johansson LF, Kulkarni P, Mei H, Neerincx P, Joeri van der Velde K, et al. (2023) Ten quick tips for building FAIR workflows. PLoS Comput Biol 19(9): e1011369. https://doi.org/10.1371/journal.pcbi.1011369
González-Beltrán A, Li P, Zhao J et al. From Peer-Reviewed to Peer-Reproduced in Scholarly Publishing: The Complementary Roles of Data Models and Workflows in Bioinformatics. PLoS One. 2015 Jul 8;10(7):e0127612. https://doi.org/10.1371/journal.pone.0127612
