GigaBlog meets Gigantum: Guest Post from Tyler Whitehouse, Dean Kleissas and Dav Clark
At GigaScience as our focus is on reproducibility rather than subjective impact, it can be challenging at times to judge this in our papers. Targeting the “bleeding edge” of data-driven research, more and more of our papers utilise technologies, such as Jupyter notebooks, Virtual Machines, and Containers such as Docker. Working these tools in to the review and publication process potentially makes it easier to assess replicability and utility, but in carrying out reproducibility case studies (see this example published in PLOS One) and getting our in-house data scientists involved in the review process, we have experienced some hurdles – one paper costing us over $1500 USD in AWS credits to recreate the workflows. As modern research is data-driven and iterative, there needs to be more practical, cost effective and efficient ways to test and review reproducibility. As we are always on the lookout for new platforms to test and integrate into our open science workflow, here, we have a guest posting from the team at Gigantum, who have developed a new open source platform that aims for better collaboration, sharing and developing reproducible research easier. Below, Tyler Whitehouse, Dean Kleissas and Dav Clark from Gigantum, highlight important issues and hurdles they have seen when attempting to make reproducible research.
Making Reproducibility Reproducible
Over the past two years our conception of what reproducibility is, and should be, has evolved into something that we can hopefully articulate with some clarity. Our perspective comes from both developing a reproducibility platform and from evaluating the practical reproducibility of various academic papers. First and foremost, it is clear to us that using re-execution of static results as the main form of verification, i.e. “reproducibility”, isn’t enough. Reproducibility should be more multi-purpose and free flowing. Basically, it should be functionally equivalent to collaboration. Additionally, it is clear to us that the academic focus on manual application of best practices as the path towards reproducibility is ineffective, and that the community should fully embrace approaches that minimize effort rather than maximize it.
In our experience, focusing reproducibility approaches on the needs of the scientific end user provides a more effective and swift path towards widespread and scalable reproducibility. This involves taking a slightly different perspective on the problem, i.e. one of creating “consumer” oriented tools and frameworks rather than one of post hoc practices and greater responsibility for researchers.

This image is under the Creative Commons License CC-BY-SA from Abstruse Goose.
Reproducible Work
The transmission of scientific knowledge and techniques is broken, but you already know this because otherwise the title wouldn’t have caught your eye. Reproducibility is the poster child for this breakdown because it is a serious problem, but also perhaps because it is tangible. What is actually wrong with the transmission of modern science is much harder to put your finger on.
We don’t want to go all Walter Benjamin on you, but scientific interpretation in the age of digital reproduction needs a deep re-think. Since that is more appropriate for a dissertation, let’s just focus on reproducibility.
Historically, reproducibility was the manual re-execution and validation of a single result, something fundamental to science going back to the 17th century. Typically the job was to recreate physical effects or observations, and manual re-execution fit well with the written descriptions of scientific results.
As digital data collection emerged in the latter half of the 20th century, expectations for reproducibility added an archival and computational component. Theoretically, data and methods were enough to understand & re-execute a work, and the ease of digital transmission made reproducibility feasible.
But did it?

Looks hard. Doesn’t look fun. This image is under the Creative Commons License CC-BY.
In the last 20 years, science continued to digitize, and complex software & intensive computation are now central to many investigations and fields. In this context, the research paper is often just a summary of activities expressed almost entirely through curated data and thousands of lines of code.
Expectations for reproducibility have changed again, and they now demand both software and data be available, while open access & open source increasingly go hand in hand. But barriers to transmission refuse to go away. Functioning software and code now depend on a techno-human tower that can fall if the slightest thing goes wrong.
Reproducible Environments
Very recently, containers (e.g. Docker & Singularity) have streamlined the creation of portable software environments. While their manual use requires significant labor and skill, they provide the most reasonable avenue to re-execution. Accordingly, expectations of reproducibility are undergoing an update and containers are a new requirement for the digital archive.
However, skill mismatches between research producer and consumer haven’t vanished yet. The ability to create and use containers presents yet another technical asymmetry that interrupts the communication & transfer of science.
Scientists will soon be subjected to a sequence of articles on best practices for containers, but the plethora of posts on Towards Data Science about “easy containers” should raise suspicion. Basic asymmetries in skill and resources between researchers still create significant difficulties in communicating and transferring scientific knowledge.
Very recently, thanks to software developers, there are user friendly frameworks, e.g. Binder, that provide streamlined re-execution environments for a broad audience. Such frameworks run in the cloud to provide ephemeral “re-execution as a service”, thus promoting wider access than previously conceivable just a short while ago.
But, we question whether “re-execution” is an actual measure of validation or reproducibility. It seems that we need something a bit more interrogative into the scientific process behind a given work.

Science needs functional tools that promote the collective effort, not a bunch of busy work. Image is the Detroit Industry Mural by Diego Rivera.
Reproducible Work Environments
When we began Gigantum, we wanted reproducibility to be part of the daily experience, not just the result of a post hoc process. Reproducible work and reproducible environments are all good, but we wanted reproducible work environments, i.e. to have the fruits of daily work be reproducible without extra effort or thought. This seemed the best way to make reproducibility reproducible.
Pushing reproducibility down into the daily experience moves it closer to real collaboration. Basically, if I can reproduce your work in my environment and you can reproduce mine in yours, then we can work together. It also means that I can interrogate your work and your process in a way that is natural to me.
So, we focused on the connection between collaboration and reproduction, and we formulated some basic requirements that we think functional reproducibility should provide:
- Integrated versioning of code, data and environment;
- Decentralized work on various resources, from laptops to the cloud;
- Editing and execution in customized environments built by the user.
Basically, we think that the line between collaboration and reproducibility should be access & permissions, and that’s all. They should be functionally equivalent. The existence proof for this is that they are functionally equivalent for two super skilled users with enough time.
To go a step further, we wanted to eliminate the effort & skill needed to create reproducible work environments and to make this capacity broadly and automatically available. So we created something that does the following.
- Focuses on the user experience and tries not to change how people work;
- Runs locally on a laptop but provides cloud access for sharing and scale;
- Automates best practices and admin tasks to save labor and add skill.
The first requirement is important for adoption because learning curves or interrupting functioning work habits won’t promote use. The second respects users’ daily lives, budgets and the occasional need for scale. The third forces the platform to promote efficiency & level out asymmetries.
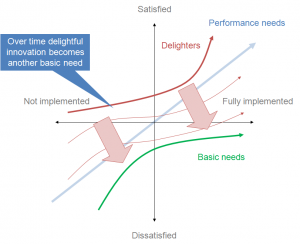
The Kano model for the user experience. Something every scientist should know. CC-BY-SA
The User Experience
A platform that satisfies the above requirements doesn’t just make reproducibility feasible. It makes it user focused.
We think it is clear that user focused approaches are best to promote real change. For example, the rapid adoption of Project Jupyter and Binder show how approaches can scale when they solve a difficult technical problem but also appeal to a broad user group.
When you think about it this way, the problem of reproducibility seems more like one of a bad user interface than a deep scientific problem. This is an oversimplification, and the issues from bad science or the inherent difficulties are deep and potentially worsening.
However, if you haven’t noticed, the business as usual approach of academia (i.e. more standards, more best practices & more training) isn’t working well. After years of urgency, it isn’t even clear if the results are mixed. Maybe they are just bad. For example, there still hasn’t been even modest adoption of best practices and results are published monthly that are not computationally reproducible. And it is for good reason.
For most researchers, the effort of manually enabling reproducibility is typically more trouble than it is worth. Basically, it boils down to bandwidth and priorities. Thinking that loosely enforced community standards can make reproducibility high enough priority that it commands sufficient bandwidth seems naive, especially if it may require potential career sacrifice.
So perhaps it is time to change the perspective from an academic one towards a more product focused one. To better promote reproducibility, people should account for the frailties of the human beings expected to enable it, and then create software that addresses those frailties. Even the champions of reproducibility sometimes have difficulty living up to their own ideals.
The best solutions make everybody on the team better, not just the most skilled. So, if people are pursuing approaches to reproducibility that don’t take a broad spectrum of end users into account, then we suspect that they may be wasting everybody’s time. And nobody can afford that.
Gigantum
At Gigantum, we develop software that increases transparency & reproducibility in science and data science. There should be no barriers to accessing science, and that is why the Gigantum Client is MIT licensed and can be run wherever you choose. Gigantum is flexible and decentralized to provide a way for anybody to easily create & share with collaborators and the rest of the world.
We made it to fit the way people actually work, that is, both locally and in the cloud. The Client runs on Docker, so using it locally is just a Docker install away, and using it on your own cloud resource is almost as easy. You can right size your resources for the problem at hand and use your own commercial cloud account for scalable compute.
Additionally, our Gigantum Cloud service also provides a storage for sharing and collaboration. You can push and pull work to and from Gigantum Cloud for access from any machine. You can also give permissioned access to collaborators so that you can share work in controlled ways. As (former) researchers, we understand the cost constraints academics have, and that is why we provide 20GB of free storage on Gigantum Cloud for storing and sharing Projects & Datasets. The service will be expanding over the next year and we welcome your input on useful and needed features in our community forum.
If you want to learn more about Gigantum try the demo and do a quick start, or install the MIT licensed software to start working locally.
This guest post has been adapted and reposted from a Medium post. For more, read the original here: https://medium.com/gigantum/making-reproducibility-reproducible-7457d656680c