GigaScience goes CC4. A handy cut out and keep guide to our licenses
We want you to use our stuff
We get many queries regarding the policies and licenses associated with our content and supporting data, and as our co-publishers BioMed Central have just announced the migration of their licenses (including ours) to the newly released version 4.0 of the creative commons CC-BY attribution license, we thought it would be a good opportunity to clarify our policies in an easier to understand manner than having to trawl through our editorial policy pages. Puneet Kishor the Science and Policy Data manager at Creative Commons gives a great overview of what the improvements of their new licenses are, and BMC has recently updated and clarified their policies for supplemental data (all CC0), but it may not be as clear what the policies are for any of our other associated materials. Are these equally open? The answer to put it simply is yes.
On top of having fully open access papers, all of the other textual content we produce such as blog posts (including this one), and the pre-publication histories of our papers (we are keen to credit our referees for the hard work they do) we want to be considered under the same CC-BY license. In general we are for anything that enables openness and transparency, so we encourage the use of pre-print servers (in fact we love them), and unlike many other publishers do not put restrictions on the use of commercial pre-print servers such as PeerJ pre-prints or BioarXiv. To summarize, our goal is to maximize the reuse of our content by making it as open and widely available as possible. While users should follow community norms such as citation, these have always We have produced a handy cut out and keep guide to summarize how these relate to journal textual content, supporting data, and the software and workflows we and our authors have hosted in the GigaScience GitHub page and GigaGalaxy servers.

We try our hardest to stick to these policies, both for infrastructure and data we produce, and of our submitting authors. The many software and data licenses out there can be complex in application and combination, we and our authors are only human, and reviewing data and software is challenging, but the aim is to push the boundaries of openness further than they currently are. There may be times we or our referees may miss something or our policies need updating or strengthening, but if the eagle eyed among you spot anything not available or contradictory in our papers or policies (many of which we have inherited from other sources) please let us know and we will fix and improve things.trailer film Inside Out 2015
While there is pushback from some sources that the lack of reproducibility and growing retraction rate of scientific papers isn’t a problem, we are at least making moves to try to address some of these issues. Many of the traditional publishers also promote non-Budapest compliant licenses under the argument that they are offering “author choice”, where in reality their motive is usually “double dipping” and monetizing their authors content further through things like reprint sales. While being freely accessible is important, being readable by machines and available for data mining is maximizing the potential of our content and making it truly open, and the BMC full-text corpus (including our papers) is available for download. We avoid non-commercial or non-derivative clauses which while allowing people to read content, restrict their reuse and people building upon them in “the commons”. These clauses allows people to look at the shoulders of giants but not stand on them, and limits their educative potential and preventing their use in platforms such as wikipedia. We’ve been pleased to see some of our useful commentaries being highlighted as useful guides for bioinformatics training.
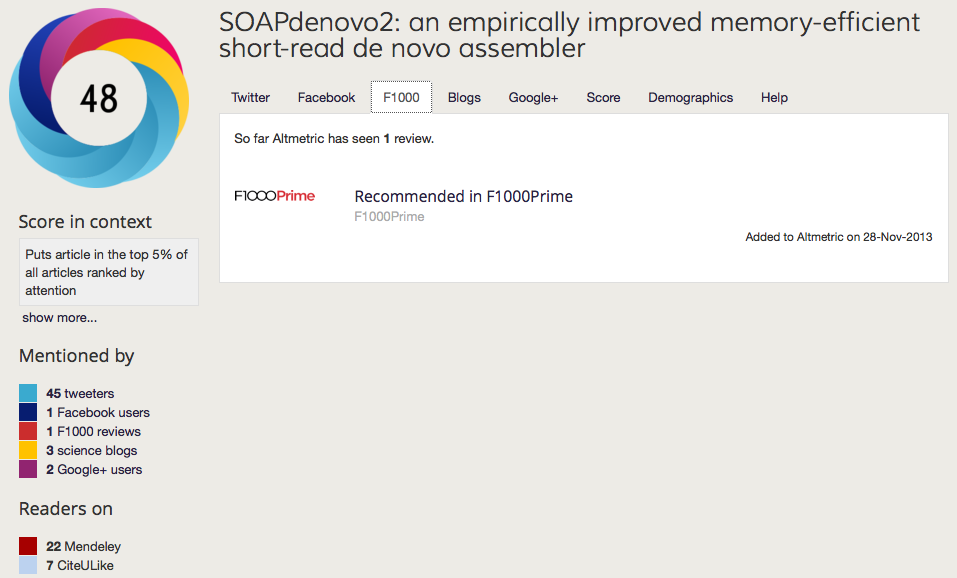
Positive examples of reuse: SOAPdenovo2
A great example of the the benefits of allowing people to build upon this open approach can been seen with our SOAPdenovo2 paper, which even though it was published less than a year ago, has already accumulated over 17,000 accesses and picked up 50 citations according to googlescholar. This has been aided by making the pipelines and 80GB of supporting data available in GigaDB, and workflows implemented in our GigaGalaxy server, and the fact that the reviews and code are available has allowed other to use it as an example of how to review software, post-publication peer-review through bloggers picking the modules and code apart in blogs and wikis, and over 18,000 downloads in sourceforge. It has also been one of our most popular articles on social media, attaining high altmetric score, and being our first article positively reviewed by F1000 Prime (as highlighted below).