Publish Peer-Reviewed Protocol Papers with Protocols.io and GigaByte
This week we are pleased to announce and highlight new updated additions to our long-running collaboration with protocols.io, linked to our first paper with a peer-reviewed protocol featuring a “Peer-reviewed method” badge on protocols.io. Alongside the addition of new functionality in protocols.io to submit protocols as method papers to GigaByte. The new GigaByte paper presents a new method expanding the alphabet and information density of DNA-data storage using methylated DNA-nucleotides, and is a great example to showcase these new features. The paper including a “wet-lab” method for experimental validation of this new “transcoding” DNA-storage scheme, alongside ”dry-lab” in-silico code that describe, encode and simulate the encoding of data in synthetic DNA using these transcoding rules. The computational methods hosted in GitHub (with snapshots in GigaDB), and the detailed “wet” protocols hosted in protocols.io.

A Publishers Protocol for Protocol Publishing
As an Open Science publisher we have been early and keen adopters of a number of third-party tools that aid reproducibility, interactivity and transparency of research, and working with our friends at protocols.io has been a particularly long-running and fruitful collaboration. Since publishing our first papers featuring the integration of protocols.io protocols in June 2016, to date GigaScience and GigaByte journals have helped contribute 144 protocols to our GigaScience Press workspace. Helping us go beyond presenting methods in brief paragraphs or supplemental downloadable PDF files that are not properly scrutinized or easily findable, to bring them center stage as first-class objects of research. The protocols.io platform enabling researchers to submit their methods in a standard format, with no space limitations, that can be directly linked to any article simply through a citable DOIs. These can also be searched online and versioned for adaptations for future work. Not only does this allow the research community easy access to detailed methods, it also means authors don’t have to continually rewrite methods for every paper that uses them. On top of the increased discoverability and permanence it allows these protocols to join the now more than 20,000 other public protocols in a single resource where a broad community of researchers can help improve methods through new versions, forks, and discussions.
Over the last 9 years how we’ve worked with protocols has evolved, starting with members of the GigaScience Press Curation and Editorial team assisting authors in getting their protocols into protocols.io. As well as the Editors taking great care to make sure protocols.io citations are correctly cited in the references (following the Data Citation Principles) to provide credit and enable tracking through citation databases. We’ve worked with the protocols.io team and others to add some commonly used genomics protocols (such as the many RNA-extraction protocols of the 1KP project) in our workspace that have been re-used by the many genome papers we publish. The launch of our GigaByte journal in September 2020 has allowed even further integration, as our (ALPSP award winning) XML-first publishing platform developed by River Valley Technologies enables more interactivity in our papers, and protocols.io protocols can be directly embedded in the methods sections of our papers. The GigaScience Press team now encouraging and assisting the authors in forming “Collections” of protocols so a single top level protocol is presented rather than the many individual protocols, particularly for studies collecting many new and previously published protocols together.
Publishing Peer-Reviewed Protocol Papers
The new features showcased this week takes things a step further, with users of protocols.io now directed to submit protocols for peer review through a “Peer Review Options” dropdown that includes GigaByte as an example of a journal that offers peer-reviewed protocol papers. Previously we would send authors in our journals to protocols.io to submit their protocols, but now creators of detailed and highly re-usable protocols can be directed in the opposite direction to us alongside other journals that publish peer-reviewed protocol papers. GigaByte has traditionally published computational methods as “Technical Release” articles (see the author instructions), but this article type is also appropriate for lab-based methods as long as they are detailed and meet our scope (i.e. are experimental methods involved in the production, analysis or handling of large-scale research data).
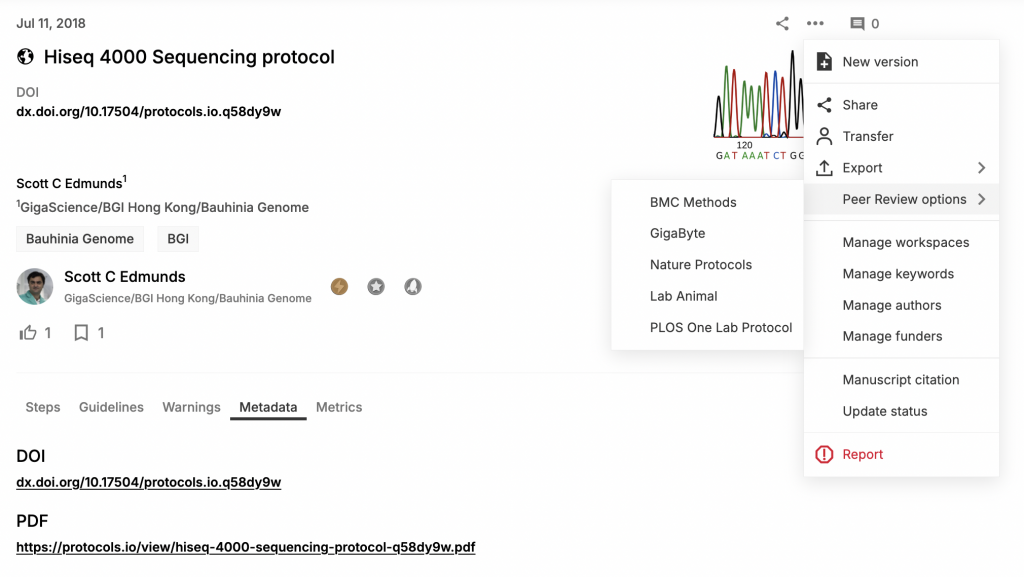
The line between computational and lab methods has increasingly blurred, and protocols.io has also got better at handling computational methods (see this handy video on how they support bioinformatics). Our recent GigaByte paper from the International Cannabis Genomics Research Consortium included very detailed protocols on how to setup a multi-omics data portal on the open-source Tripal platform. In this weeks new example we have showcased a paper with a detailed lab-protocol, and going forward please think about sending your lab-based methods as Technical Release papers in GigaByte. To assist this process protocols.io have provided the following author guidelines on publishing protocols in methods journals:
Availability of protocols to reviewers and other researchers
Publishing research protocols on protocols.io is encouraged; a digital object identifier (DOI) can be assigned to the protocol as a permanent link to the method, which can then be dynamically updated. To include a link to a protocol in your manuscript:
1) Add your step-by-step protocol on protocols.io and then follow the instructions here: https://www.protocols.io/help/new-methods-development/publish-articles.
2) Select either ‘Publish’ or ‘Reserve DOI’ to generate the link(s) to put in your manuscript (it is recommended to publish and make fully available but appreciate that some will want their protocol private initially).
3) Provide editor and reviewer access by Including the DOI (and private) link(s) and a reference for your protocol in the relevant section of your manuscript using the format provided by protocols.io: http://dx.doi.org/10.17504/protocols.io.xxxxxxx (where xxxxxxx is the unique DOI).
4) Add a citation to your protocol in the references section of your manuscript. Citation format should be according to the guidelines of the journal, but should include the following fields:
Authors. Protocol Name. protocols.io
https://dx.doi.org/10.17504/protocols.io.xxxxxxx (where xxxxxxx is the unique DOI)5) If the DOI was reserved, you’ll need to return to your protocol and select ‘Publish’ to make the protocol publicly available. Remember to also remove any private links from your manuscript and only leave the DOI.
When protocols are peer-reviewed we now have the ability to highlight this with a “Peer-reviewed method” badge on protocols.io, and now we have this feature we will be sending associated protocols to our peer reviewers to see if they have any additional comments and feedback on them. Adding value, scrutiny and improving the reproducibility of published research, and providing the credit to the reviewers and authors for these efforts.
If you’d like to keep up with our latest published and peer-reviewed protocols please follow our protocols.io workspace:
https://www.protocols.io/workspaces/gigascience-press
References
Liu D et al., A practical DNA data storage using an expanded alphabet introducing 5-methylcytosine, GigaByte, 2025 https://doi.org/10.46471/gigabyte.147
Liu D et al., Experimental validation for transcoding scheme name R+ in DNA data storage. Protocols.io. 2024; https://dx.doi.org/10.17504/protocols.io.q26g7mr78gwz/v1
Edmunds SC and Goodman L, GigaByte: Publishing at the Speed of Research, Gigabyte, 2020 https://doi.org/10.46471/gigabyte.1