Some sea cucumbers like it hot
We start the new year with news from the deep, published in GigaScience: The genome of a sea cucumber, collected at a depth of 2400 m during a submarine trip to a hydrothermal vent, helps scientists to understand how marine animals can survive in extreme conditions.
Hydrothermal vents are an unlikely environment for animals to flourish. Temperatures change dramatically, from really cold to boiling hot, and the chemistry near an active vent is something else altogether: acidic pH, sulfur and methane are the typical combination. Not to mention the high hydrostatic pressure and the darkness of the deep sea.
The research team of Haibin Zhang from the Institute of Deep-sea Science
and Engineering (Chinese Academy of Sciences) have now sequenced the full genome of a particularly unusual inhabitant of the hydrothermal vent environment: the sea cucumber Chiridota heheva.
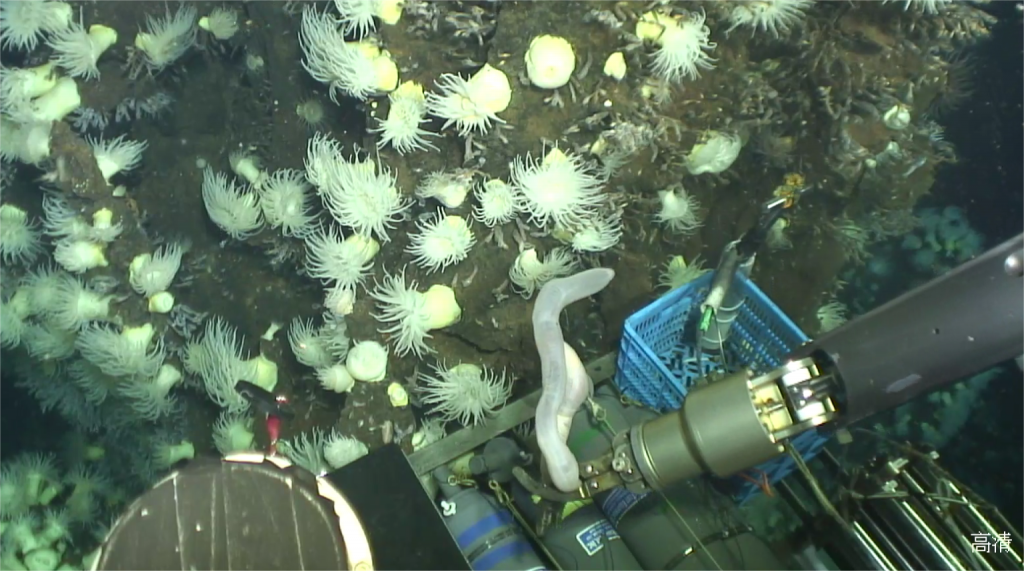
Unique life forms
Organisms found at hydrothermal vents are among the most unique life forms on the planet. They evolved special adaptations to survive and procreate under these harsh conditions. For example, many microbes employ special metabolic functions to deal with the abundance of sulfur and iron, and to withstand the enormous heat near the vent. In addition to microbes, there are even multicellular and higher order organisms that have adapted to the hydrothermal vent conditions, including various species of worms, snails, crabs and shrimp.
In 2019, a Chinese deep sea expedition with the manned research submarine “Shenhaiyongshi” collected a specimen of the sea cucumber C. hehevae at the bottom of the Indian Ocean, at the Kairei vent field at a depth of 2,428 meters. The water around the Kairei vent is rich in dissolved iron, adding to the already harsh conditions.
Sea cucumbers are echinoderms, and as such related to sea urchins and sea stars – a group of animals with highly unusual body plans. They are found on sea floors all over the world, where they devour detritus and use their tentacle to explore the sediment. While other high-quality genomes of sea cucumbers are available, the work now presented in GigaScience is the first high-quality genome of a sea cucumber specimen collected at a hydrothermal vent.
Expanded sea cucumber gene families
Initial comparative genome analyses indicate that several gene families are expanded in this sea cucumber, meaning that the species has a higher repertoire of specific sets of genes than related species.
Specifically, gene family analyses revealed that 450 gene families expanded and 6 contracted in C. heheva, and that some of the expanded gene families have roles in iron binding and transport – indications that these genomic changes may be interpreted as adaptations to the iron-rich environment.
Another group of genes that show indication of adaptation are genes related to DNA repair, and again this can be interpreted as adaptation to the harsh environment. While the details will need to be studied in more detail, the genomic data that’s now available are a valuable resource for further studies on both, sea cucumbers and the unique vent fauna.
References:
A high-quality chromosomal genome assembly of the sea cucumber Chiridota heheva and its hydrothermal adaptation
Yujin Pu, Yang Zhou, Jun Liu, Haibin Zhang
GigaScience, 2024, giad107 https://doi.org/10.1093/gigascience/giad107
Supporting data for “A High-Quality Chromosomal Genome Assembly of the Sea Cucumber Chiridota heheva and Its Hydrothermal Adaptation.”
Yujin Pu, Yang Zhou, Jun Liu, Haibin Zhang
GigaScience Database. 2023 https://doi.org/10.5524/102481