The Early Earthworm Catches on to Full Data Release
New research and data published in GigaScience and PLOS ONE provides complete open access to detailed 3D images of earthworms
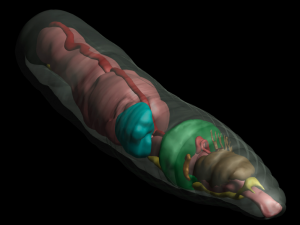 To quote the American cartoonist Gary Larson: all things play a role in nature, even the lowly worm—but perhaps never in such a visually stunning way as that presented in two papers published last week in GigaScience and PLOS ONE. The work and data presented here provide the first-ever comparative study of earthworm morphology and anatomy using a 3D non-invasive imaging technique called micro-computed tomography (or microCT), which digitizes worm structures. This opens the possibility of scanning millions of specimens from museum collections, including extinct species, all of which is important given that the earthworm is both a benefit and a bane to ecosystems. Being open access, the stunning high-resolution images, videos and interactive models also can be used as teaching aids and make fantastic resources for understanding worm anatomy (see the screenshot from the PLOS supporting data).
To quote the American cartoonist Gary Larson: all things play a role in nature, even the lowly worm—but perhaps never in such a visually stunning way as that presented in two papers published last week in GigaScience and PLOS ONE. The work and data presented here provide the first-ever comparative study of earthworm morphology and anatomy using a 3D non-invasive imaging technique called micro-computed tomography (or microCT), which digitizes worm structures. This opens the possibility of scanning millions of specimens from museum collections, including extinct species, all of which is important given that the earthworm is both a benefit and a bane to ecosystems. Being open access, the stunning high-resolution images, videos and interactive models also can be used as teaching aids and make fantastic resources for understanding worm anatomy (see the screenshot from the PLOS supporting data).
From digging in the garden to fishing to playing with worm farms at school, everyone has become aware of the importance of earthworms in maintaining ecosystems and soil quality, but few realize that many earthworm species are invasive and can be a serious threat to their new environment. More difficult yet is distinguishing one worm from another, and with thousands of known species around the globe, differentiating them and resolving their taxonomy is an enormous task. The difficulties in dissecting small, soft-bodied organisms, as well as having access to valuable and rare museum specimens, can make doing so nearly insurmountable. However, the research published by Dr. Rosa Fernández and her colleagues from Harvard University as well as Dr. Alexander Ziegler from Ziegler Biosolutions provides the much-needed tools to carry out such analyses in a non-destructive way and at a significantly quicker pace. Their work combines two- and three-dimensional imaging techniques and shows that commonly used morphological features for defining earthworm taxonomy can now be assessed without the need for dissection. The researchers were able to view the sediment inside the animal in 3D, as well as the internal organs in context with each other. They also compared fresh samples with preserved museum specimens, going as far back as 1945, and revealed that this digital imaging technique is suitable for the analysis of rare museum specimens.
On top of releasing beautiful high-resolution videos allowing non-destructive 3D real-time dissection of earthworms, the authors made supporting raw and derived data available in our GigaDB repository, enabling future data mining by researchers, the development of new tools, as well as provision of increased transparency and confidence in the research. Lead and corresponding authors on the papers, Dr. Rosa Fernández and Dr. Alexander Ziegler say that they would be delighted if their study “will serve as a catalyst for future high-throughput studies of zoological specimens and will also boost enforced data deposition practices in biodiversity research”, opening new possibilities for computer-based interactive exploration and study of museum specimens around the world.
Making the data openly available is especially unique because, although open access to supporting data is the norm in some areas of research, for the morphology and taxonomy community this is much less common and, in practice, more difficult. Such issues have led to community concerns that data deposition policies adopted by journals such GigaScience and PLOS may not be able to deal with large and complex datasets, such as these large stacks of imaging files. Thus, the data release effort by the authors and by GigaScience and PLOS ONE working together constitutes a major step forward in data transparency, making use of the online database MCZbase and the big data capabilities of our GigaDB repository. This follows up our recent release of similar centipede microCT data in GigaDB last year (see “the cyber centipede”), demonstrating the utility of the platform for large scale imaging datasets that otherwise would not have a home. Despite some of the folders being close to 10 GB in size, the data reviewers were able to retrieve each of those in as little as half an hour using our high-speed Aspera internet connection (for more on that see our previous post).
As with all our research, peer-review was carried out transparently and is viewable from the pre-publication history of the article. Dr. Sarah Faulwetter of the Hellenic Centre for Marine Research, who served as a reviewer, felt that the level of data availability brings taxonomy into a new age, saying, “Instead of having to go through the lengthy process of obtaining the physical specimen from a museum, I can now download a fairly accurate representation from the web.” With the goal to maximise future reuse of the data, Dr. Faulwetter further comments: “The authors have also made the raw data (the CT projection images) available – this gives me as a researcher the additional possibility to reconstruct the 3D dataset again with different parameters, allowing me to focus exactly on the features I need.”
As the British natural scientist Charles Darwin said of the humble earthworm in the late 1800s, “It may be doubted whether there are many other animals which have played so important a part in the history of the world, as have these lowly organized creatures”, but, today, how they are organized has never been seen so clearly or been so easily viewed.
References
1. Lenihan, J; Kvist, S; Fernández, R; Giribet, G; Ziegler, A (2014) A dataset comprising four micro-computed tomography scans of freshly fixed and museum earthworm specimens. GigaScience, 3:6 http://dx.doi.org/10.1186/2047-217X-3-6
2. Lenihan, J; Kvist, S; Fernández, R; Giribet, G; Ziegler, A (2014): MicroCT scans of freshly fixed and museum earthworm specimens. GigaScience Database. http://dx.doi.org/10.5524/100092
3. Fernández, R; Kvist, S; Lenihan J, Giribet G, Ziegler A (2014) Sine systemate chaos? A versatile tool for earthworm taxonomy: non-destructive imaging of freshly fixed and museum specimens using micro-computed tomography PLOS ONE 9 (5) e96617 doi:10.1371/journal.pone.0096617
Recent comments
Comments are closed.
[…] of biology, the size and scope of where the field of genomics is now. Following closely on from the large worm imaging dataset we also published as a Data Note this month, this paper further shows the utility of our […]
[…] of biology, the size and scope of where the field of genomics is now. Following closely on from the large worm imaging dataset we also published as a Data Note this month, this paper further shows the utility of our […]