The genome of a shape shifting butterfly
A high coverage, high quality genome sequence of the butterfly Bicyclus anynana is published today in GigaScience. B. anynana, the squinting bush brown, is a fascinating model species that can modify its morphology in response to environmental clues. The 475 Mb genome assembly, achieved by combining traditional Illumina and long read PacBio data, encodes 22,642 predicted protein-coding genes and will be a resource for a wide range of studies, from developmental biology to population genomics. The high quality assembly by Reuben Nowell et al. is the product of a collaboration of Mark Blaxter’s genomics group at the University of Edinburgh with an international team of lepidopterists. It further demonstrates that long-read derived genomes are increasingly becoming the standard in genomics, as we described in a recent GigaBlog post.

B. anynana
credit: William H. Piel and Antónia Monteiro
When Paul Brakefield (an author on this paper) collected 81 gravid females of squinting bush brown butterflies during a 1988 field trip to Malawi and brought them back to his former lab in Leiden, the Netherlands, a new model organism took off: Bicyclus anynana was now available for large-scale genetic investigation in controlled, lab-reared stocks. The species has some practical advantages. B. anynana butterflies happily breed in captivity. They are small enough to be kept in large numbers, but big enough to perform microsurgery experiments, for example to interfere with wing development.
However, the reason why this tropical butterfly is such an intriguing species for ecologists, developmental and evolutionary biologists alike reaches beyond practicalities. The squinted bush brown has a unique biology. It is a shapeshifter – in scientists‘ jargon, it displays „seasonal polyphenism“. During the short, hot rainy season biologists catch mostly animals that display conspicuous markings on the wing, so-called eyespots. During the long, cold dry season however, individuals of the very same species, with the same genetic background, come with a different and much more boring look: They are uniformly brownish.
A different shape for each season

Two ways ahead: a B. anynana egg. credit:
The decision whether an individual butterfly develops into an eyespot-decorated beauty or into a well camouflaged brownish thing is made very early, during larval development. In other words, a B. anynana egg has two alternative, very different futures ahead – and the decision which route is taken is not triggered by genes, but by environmental clues. In particular, declining temperatures initiate the development of the dry season form. This switch mechanism is evolution‘s answer to two different selection pressures. In dry season, it is an advantage to keep a low profile to evade vertebrate predators, whereas in wet season ample mating opportunities – and the need to deflect predator attacks with the help of eyespots – outweigh the risk of being detected and eaten by the wrong guy.
Physiologically speaking, ecdysteroid hormones are the key players and turn on the transcription of developmental tool kit genes, leading to development of the seasonal form.
To understand this switch mechanism, evolutionary and developmental biologists, ecologists, geneticists and genomics folk have to work together. Although the cues for triggering one of the two forms are environmental, the switch itself, the capacity to choose between and execute two different developmental programs, is encoded in the genome. Paul Brakefield and colleagues have already achieved insights into the genetic architecture of this polyphenism, but the high quality genome assembly now published in GigaScience (with data in GigaDB here) will push these efforts to a new level. Hopefully, this will lead to a fuller picture of how evolution tinkered with the B. anynana genome, such that it can orchestrate two widely different, alternative outcomes.
A boost for insect genomics
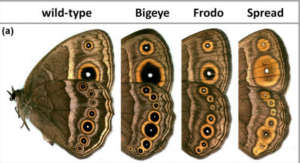
Mutants with altered eyespots
credit: SV Saenko et al, BMC Biol 8:111, CC-BY
B. anynana encodes more genes than other sequenced lepidopterans, with ~3,250 more genes than the diamondback moth P. xylostella, and ~10,400 more than the swallowtail P. polytes. It is tempting to attribute the apparently high number of genes to the developmental plasticity and alternative seasonal forms in B. anynana. However, it remains to be determined whether the number of genes predicted in B. anynana is a function of its larger genome size, the unusual life history, or if further research and curation will reduce the number of inferred genes.
During the last couple of years, a large number of insects had their genomes sequenced – a massive boost for projects such as i5K and 1KITE, and in general for all efforts to figure out the complicated phylogenomic relationships of arthropods. At GigaScience, we are happy to do our bit to advance these goals. For example, we recently published Data Notes on the genome sequences of the Antarctic winged midge and of the sweetpotato whitefly.
The unusual seasonal polyphenism of the squinted bush brown is just one of several interesting questions that can now be explored with the help of a high quality genome sequence. For example, developmental biologists are interested in the markings on butterfly wings (even going as far as modifying these with genome editing), and ecologists are fascinated by the massive radiation of Nymphalidae, the family to which B. anynana belongs.
Whether it‘s dry or wet season, butterfly researchers will be busy.
References
-
- Nowell, R, W; Elsworth, B; Oostra, V; Zwaan, B, J; Wheat, C, W; Saastamoinen, M; Saccheri, I, J; van’t-Hof, A, E; Wasik, B, R; Connahs, H; Aslam, M, L; Kumar, S; Challis, R, J; Monteiro, A; Brakefield, P, M; Blaxter, M (2017): A high-coverage draft genome of the mycalesine butterfly Bicyclus anynana. https://doi.org/10.1093/gigascience/gix035
- Nowell, R, W; Elsworth, B; Oostra, V; Zwaan, B, J; Wheat, C, W; Saastamoinen, M; Saccheri, I, J; van’t-Hof, A, E; Wasik, B, R; Connahs, H; Aslam, M, L; Kumar, S; Challis, R, J; Monteiro, A; Brakefield, P, M; Blaxter, M (2017): Supporting data for “A high-coverage draft genome of the mycalesine butterfly Bicyclus anynana” GigaScience Database. http://dx.doi.org/10.5524/100280
- Brakefield PM , Beldade P and and Zwaan BJ (2009) The African Butterfly Bicyclus anynana: A Model for Evolutionary Genetics and Evolutionary Developmental Biology doi:10.1101/pdb.emo122