We Wish You a GigaChristmas: A 2015 Wrap-Up
Last Christmas we gave you our heart; okay forget George Michael – we gave you beautiful imaging data sets, Virtual Machines, BYO data parties, GigaGitHub, more open peer review plus much more. However, in 2015 GigaScience has delivered anything but less, with more technical developments and exemplar papers published – GigaScience continues to push the boundaries of reproducible research and innovative publishing of ALL research objects (article + data + code + workflows + VMs/Docker). With 2016 rapidly approaching, we thought we would give you a summary of our recent technical developments over the year, including published examples that showcase them.
More Technical Publishing
Our repository, GigaDB, recently underwent a technical service representing almost a year of hard work, with many efforts behind the pretty façade. The main feature changes were recently highlighted in a blog by our Lead Curator, Chris Hunter, which include changes to data structure, submission wizard, a much improved search function and results display, and the highlighting of our advisory board and FAQ information. In addition, Hypothes.is web annotation tools have been implemented in GigaDB – meaning anyone can now annotate datasets hosted in GigaDB, which fosters more reproducible science and transparency. Our GigaGalaxy computational workflow publishing platform has also had an under-the-hood tune up, with a migration to new servers (and http://gigagalaxy.net/ URL) greatly speeding up access and improving stability. The last few months has also seen a lot of new Galaxy papers published, so follow our Galaxy series page for the latest.
GigaScience was also pleased to recently announce our partnership with the Hypothes.is “Annotate All Knowledge” initiative which, along with 40 other scholarly publishers, platforms and technology partners, aims to build an open conversation layer over all knowledge. You can read more about this initiative in our Q&A with Hypothes.is Director of BioSciences, Maryann Martone.
Earlier in 2015, Chris Hunter and “Jesse” Xiao Si Zhe from our curation team, attended the Biocuration 2015 meeting in Beijing, where they presented posters and participated in a workshop on “Crowd/community curation: challenges & credit attribution”, chaired by our Ed Board Member Henning Hermjakob. Read more highlights from this meeting here, and watch this space for news on some exciting developments to come at the 2016 meeting in Geneva.
Ain’t no party like a Birthday, Bioinformatics and Metabolomics Party
Being regular attendees of ISMB (Intelligent Systems for Molecular Biology) meeting, we have some history – with having launched at the 2012 meeting in Long Beach, as well as organising workshops the previous two years (read more here and here). It was great to be able to celebrate our 3rd birthday in Dublin where we hosted a celebration at the Ferryman Pub and caught up with several Editorial Board members, authors and friends. You can read more about the highlights from this meeting here. While in Dublin we also attended BOSC (Bioinformatics Open Source Conference) where Alejandra González-Beltrán from the ISA-team presented results from our case study published in PLOS One, which we worked on with members of the Research Object, Nanopublication and ISA communities. This follows from the workshops we co-organised over the last few years, particularly 2013’s “What Bioinformaticians need to know about digital publishing beyond the PDF”.
GigaScience also throws a good (BYO data) party, and this year we’re pleased to announce that we have obtained our second BBSRC UK-China collaboration grant to further support the sharing of data and analyses in metabolomics – this is in partnership with the EBI, The Universities of Birmingham, Manchester and Oxford, as well as the Sainsbury Laboratory, TGAC and BGI. Our first collaboration grant helped us organise our “Bring your own data” hackathon in China last year (see our write-up), and also follows collaborations (and NERC funding) to work on open metabolomics workflows with the University of Birmingham. The outputs of this, and a growing number of metabolomics paper are starting to be published in the journal. Read more about our metabolomics data and training efforts in our blog here.
I want my GigaTV
 On top of ISMB this has been another packed year of meetings and workshops, with us participating in and supporting the first Galaxy Community Conference Data Wrangling hackathon, the ISA “Hack-the-Spec” hackathon in Oxford, the “hack the human genome” event in Hong Kong, and the Community Genomes workshop at BGI’s ICG10 conference in Shenzhen. If you missed them you can luckily watch many of these online, with the public summary of the genome hacking event on MakerBay TV, and our GigaTV youtube channel hosting the Community Genomes event (see also the blog). On the subject of Community Genomes, Scott and Rob from the GigaScience team have helped kickstart their first citizen science crowdfunding project with BauhiniaGenome. Taking the message onto the front pages of the Sunday papers and on the radio, check out their crowdfunding video and there is still time to support the project and pick up some Christmas prezzie perks before the deadline next week.
On top of ISMB this has been another packed year of meetings and workshops, with us participating in and supporting the first Galaxy Community Conference Data Wrangling hackathon, the ISA “Hack-the-Spec” hackathon in Oxford, the “hack the human genome” event in Hong Kong, and the Community Genomes workshop at BGI’s ICG10 conference in Shenzhen. If you missed them you can luckily watch many of these online, with the public summary of the genome hacking event on MakerBay TV, and our GigaTV youtube channel hosting the Community Genomes event (see also the blog). On the subject of Community Genomes, Scott and Rob from the GigaScience team have helped kickstart their first citizen science crowdfunding project with BauhiniaGenome. Taking the message onto the front pages of the Sunday papers and on the radio, check out their crowdfunding video and there is still time to support the project and pick up some Christmas prezzie perks before the deadline next week.
More credit to reviewers and now, authors
As you all know by now, GigaScience has always liked giving credit where credit is due, beginning with peer reviewers with our open peer review policy, followed by the addition of our open reviews on Publons – you may have thought “how much credit can a journal give back?” Well, once again, we’ve taken it a step further – to celebrate peer review week we announced the assignment of our DOIs to reviews in Publons, which means our reviews are searchable on and via the many sources that index these, such as DataCite metadata search, the Thomson Reuters Data Citation Index, and the OSF Share registry.
We also think our authors deserve more credit too, so we decided to put credit back into the hands of researchers. In late September we launched a project that provides a new way to give all authors credit for their work with a hope it will improve collaboration, transparency and better research, through Author Contributorship Badges. The badges are based on a taxonomy around contributorship developed by the Wellcome Trust, MIT, Digital Science, and others in partnership with CASRAI (Consortia Advancing Standards in Research Administration), National Information Standards Organization (NISO), and the research community. Led by Amye Kennall, Associate Publisher at BioMed Central, we have worked with Mozilla Science Lab, ORCiD, and Ubiquity Press to develop an open source system for generating transparent, validated data around contributorship through the Mozilla Open Badge Framework. Our GWATCH Technical Note is an example of this badging at work that nicely demonstrates the different roles of each author listed in the paper.
September we launched a project that provides a new way to give all authors credit for their work with a hope it will improve collaboration, transparency and better research, through Author Contributorship Badges. The badges are based on a taxonomy around contributorship developed by the Wellcome Trust, MIT, Digital Science, and others in partnership with CASRAI (Consortia Advancing Standards in Research Administration), National Information Standards Organization (NISO), and the research community. Led by Amye Kennall, Associate Publisher at BioMed Central, we have worked with Mozilla Science Lab, ORCiD, and Ubiquity Press to develop an open source system for generating transparent, validated data around contributorship through the Mozilla Open Badge Framework. Our GWATCH Technical Note is an example of this badging at work that nicely demonstrates the different roles of each author listed in the paper.
Publishing All Research Objects
Since we not only publish data, but also research, code, workflows and virtual machines and Containers, it’s only fitting that we highlight exemplar articles that showcase the variety of research objects that we publish. With regards to virtual containers, such as Docker, our Galaxy series has seen the addition of two new papers utilizing this technology. Last August we published a Technical Note where the sequence comparison tool suite BLAST+ was wrapped for easier use with the Galaxy workflow system, and was also made available in MyExperiment and as a Docker Image. Another paper published early this month describes how data from the OpenLifeData2SADI project is merged with other SADI services using Galaxy and Docker, further enhancing reproducibility and sharing of complex data retrieval and analysis workflows. Docker seems to be a technology of its time, and the bioboxes community published a call-to-arms with us trying to define standards for interoperability in container publishing. Further demonstrating it utility, we also published a Data Note that perfectly demonstrated the potential of Docker to tackle problems of un-reproducible research, with all the open data and code published in a containerized form the reviewers managed to exactly recreate the details presented in the paper. This research also allowed the authors to tackle climate change through the characterization of complex communities of micro-organisms in a biogas plant that generates heat and power from maize silage and pig manure. You can read more about this state-of-the art method of publishing research in our blog.
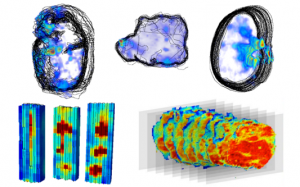 2014 was the year we gave you 3D images of earthworms and MRI data of blood perfusion through the myocardium presented in a virtual machine; this year, we have published beautiful brooding brittle star images, in coordination with supporting research published in the African Journal of Marine Science. We also published a Q&A blog with lead author, Jannes Landschoff. In addition, we published a data note showcasing benchmark 3D-MALDI imaging datasets and algorithms collected from five samples using two types of 3D imaging MS – a method challenged by the lack of publicly available benchmark datasets and tools.
2014 was the year we gave you 3D images of earthworms and MRI data of blood perfusion through the myocardium presented in a virtual machine; this year, we have published beautiful brooding brittle star images, in coordination with supporting research published in the African Journal of Marine Science. We also published a Q&A blog with lead author, Jannes Landschoff. In addition, we published a data note showcasing benchmark 3D-MALDI imaging datasets and algorithms collected from five samples using two types of 3D imaging MS – a method challenged by the lack of publicly available benchmark datasets and tools.
More Big Data
Thrown into the mix is another extremely large dataset representing 12Tb of genome sequence data and assemblies from the Canadian Cattle Genome Project. This is the second largest dataset we have published after the 13Tb of genome sequence data from the 3,000 Rice Genome project released last year on World Hunger Day.
From a different field, we published a Database (LAGOS) aimed at connecting lake temporal and satellite data, as part of our “Big Data in Ecology” series. Created by Pat Sorrano and coauthors, LAGOS is a multi-scaled geospatial temporal ecology database bringing together the many different, scattered and disparate data on ecosystems – its open-science perspective helping address the challenges of reproducibility. Pat gives us more insight into the challenges of large-scale data integration in her Q&A blog here. With just a few of our highlights from 2015, our Team can only expect more exciting things to come in 2016, so watch this space!