Sequencing genomes across the planet at Biodiversity Genomics 2021

Genomics is a powerful technology that helps us understand the Tree of Life. Biodiversity Genomics 2021 was a virtual conference that took place on 27th September-1st October 2021 that demonstrated how genomics can inform conservation and food security, and can additionally help us understand evolutionary novelties such as symbiosis. Building on the accomplishments of the Genome 10K community and the Vertebrate Genomes Project, this conference also highlights the current status of the Earth BioGenome Project. Formerly the G10K meeting, GigaScience are regular attendees of this conference (see GigaBlog of Biodiversity Genomics 2020 and Scott’s talk in the Annotation & Databases track last year) and GigaScience Data Scientist Chris Armit reports on some of this year’s highlights from sequencing the Tree of Life.
Update on the Earth BioGenome Project
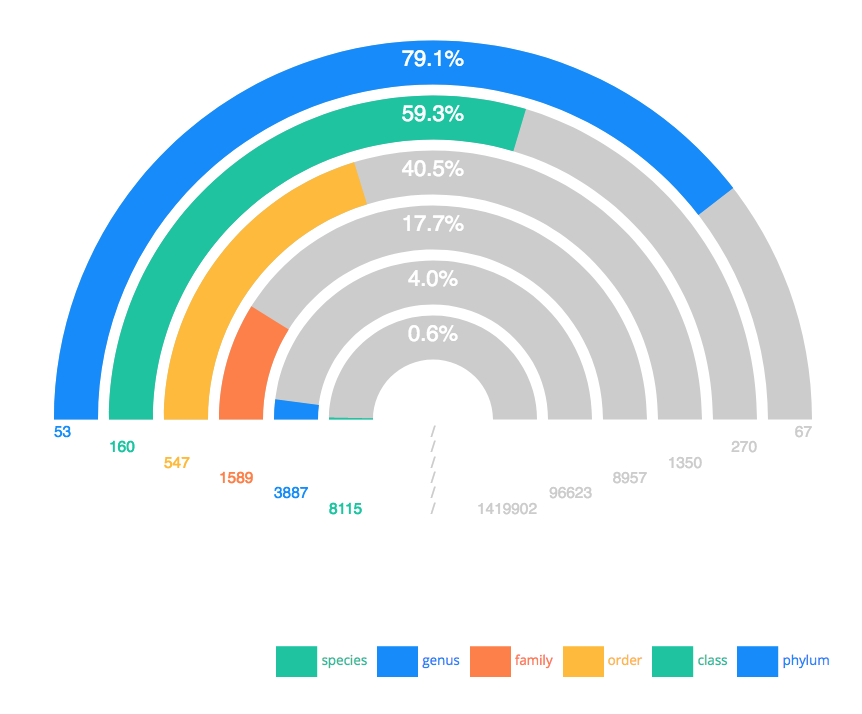
Harris Lewin (UC Davis), in his plenary talk entitled “EBP 2021: More than a Moonshot” highlighted the goals of the Earth BioGenome Project. As Harris explained, “The Earth BioGenome Project is a confederated international network-of-networks that has the common goal of sequencing and annotating the genomes of all 1.8 million known species of eukaryotes in 10 years”. Harris provided an overview of the accomplishments of the EBP and the global status of this immense research effort is that 0.6% of all known eukaryotic species have now been sequenced, with at least one representative species from 17.7% phylogenetic families, and 59.3% phylogenetic classes (i.e. amphibia, reptilia, mammalia, aves, etc).
The pipeline from sequencing to complete genome assembly involves multiple steps, and this process is underway with the generation of contig-level assembly being followed by a scaffold-level genome assembly, a chromosome-level genome assembly, and a complete genome assembly with annotation as the end product. As Harris further explained, only 15.4% of the eukaryotic genome assemblies are chromosome-level, and so currently the majority of the sequenced genomes are still at the scaffold-level or contig-level, and continued effort is needed to meet the 10-year goal.
BGI Co-Founder Huanming Yang followed on from Harris and was swift to point out that “the culture of collaboration has reshaped global sciences” and offered his continuing support “towards sequencing every living thing on the planet”.
How the Caecilian lost its Limbs
“Cursed are you above all livestock
and all wild animals!
You will crawl on your belly
and you will eat dust
all the days of your life.”
Genesis 3:14
There have been many tales as to how the reptilian serpent lost its limbs. There exists a lesser-known genus of serpentine amphibians, known as Caecilia, that are more closely related to frogs and salamanders but that have also lost their limbs through their adaption to a fossorial (burrowing) lifestyle. A question for comparative genomics is whether the mechanism by which limbless caecilians came to be is convergent with the evolutionary mechanism that has been postulated for snakes. Marcela Uliano da Silva (Tree of Life Programme, Wellcome Sanger Institute) and Vladimir Ovchinnikov (University of Nottingham) offered a genomics perspective on this question in their talk entitled “Three high quality Caecilian genomes reveal a common route to limblessness in vertebrates”. As Marcela explained, high-quality chromosome-level genome assemblies were generated for Geotrypetetes seraphini, Microcaecilia unicolor, and Rhinatrema bivittatum, and these were all found to be very large genomes, as is typical for amphibians, with a range from 3.7Gb to 5.3Gb.
To compare the genetic basis of limblessness between caecilians and snakes, Vladimir Ovchinnikov reported on the status of an enhancer known as ZRS, which is a long-range enhancer that is solely responsible for Shh expression in the limb (for more on ZRS enhancer see GigaBlog of Cell Bio Virtual 2020). As Vladimir explained, progressive loss of function of the ZRS limb enhancer has been observed during snake evolution. However, when one looks for the ZRS enhancer in caecilian genomes, it has completely disappeared and cannot be found by a BLAST search. The implication is that limblessness has evolved independently in caecilian and snakes, but is underpinned by changes to the ZRS enhancer element. This is a fascinating example of convergent evolution, and Vladimir suggests that the obliteration of the ZRS sequence in caecilians may highlight that limblessness is much older in amphibia than in reptiles.

Marcela is one of our authors and has recently published the genome assembly of the tambaqui (Colossoma macropomum) in our new journal GigaByte. The tambaqui is an emblematic fish of the Amazon River Basin and is a leading species in Brazilian aquaculture and fisheries. We’ve been fans of Marcela’s “community genomics” approach to genome projects, and relating to this published the partly crowdfunded genome assembly of the invasive golden mussel Limnoperna fortunei in GigaScience. This molluscan species has aggressively invaded South American freshwaters where it has outcompeted native species and economically harmed aquaculture. These genome assemblies highlight novel ecological and evolutionary insights, and are an important resource in ensuring that aquaculture and fisheries are sustainable.
How Bats are resistant to SARS-CoV2
In the session on bat genomics, BAT1K co-director and GigaScience Editorial Board Member Emma Teeling (University College Dublin) highlighted that bats have multiple novel adaptations, including flight and echolocation, but also a unique tolerant immunity. Using genomics, Ariadna Morales of the LOEWE Centre for Translational Biodiversity Genomics offered invaluable insights as to how bats – which are a known reservoir for coronaviruses – are themselves resistant to coronavirus-related disease. As Ariadna explained, bats of the families Hipposideridae and Rhinolophidae are the main suspected reservoirs of many coronaviruses, including SARS-CoV2.
Bats possess a special immune system that allows them to live with coronaviruses, and to address the co-evolution of this phenomenon, the BAT1K consortium generated high-quality chromosome-level genome assemblies of 7 species of bat (Aselliscus stoliczkanus, Rhinolophus affinis, Rhinolophus luctus, Rhinolophus sedulus, Rhinolophus trifoliatus, Hipposideros larvatus, Doryrhina cyclops) that are considered to be potential SARS-CoV2 reservoirs. Through comparison with other mammalian species, genomic analysis highlighted 29 genes in potential SARS-CoV2 reservoir bats that are under selection or lost in these bat species. Interestingly, the proteins encoded by these genes are targeting RNA synthesis, translation and assembly of coronavirus, but not maturation and release of virion particles. From these findings, Ariadna suggests that within Hipposideridae and Rhinolophidae, ancestral lineages may have evolved adaptations to inhibit viral replication. In contrast, species-specific adaptations to novel viruses – such as SARS-CoV2 – may inhibit translation of structural proteins and virion assembly.

For more on bats, see recent GigaBlog by Hans Zauer on a genomics perspective on the feeding habits of bats.
The Biology of Bioluminescence in Cardinalfish
“Be a lamp unto yourself.”
Gautama Buddha
Alison Gould (California Academy of Sciences) delivered a fascinating talk entitled “Shedding light on symbiosis: lessons from a bioluminescent cardinalfish”. Cardinalfish of the genus Siphamia are coral reef-dwelling fish that have evolved a symbiotic relationship with luminous bacteria. This evolutionary adaptation involves a novel organ, known as the light organ, which forms from an outcropping of the developing gut. Bacterial symbionts of the genus Photobacterium that are ingested during larval development pass through the alimentary system and seed the light organ, creating a microbiome with bioluminescent properties. A notable feature of the host cardinalfish is that it releases excess symbiont bacterial cells daily with faeces, and in doing so has the curious ability to excrete ‘glowing poop’. It is thought that bioluminescence helps the cardinalfish avoid predators by providing counter-shade on moonlit nights on the reef.
To understand the genomic basis of symbiosis, Alison and colleagues generated a high-quality chromosomal-level genome assembly of the bioluminescent cardinalfish Siphamia tubifer using a combination of PacBio HiFi sequencing and Hi-C technologies. The 1.2 GB genome assembly was then compared with the genomes of a non-luminous cadinalfish Sphaeramia orbicularis and a more distant relative the mudskipper Periophthalmus magnuspinnatus (order Gobiiformes), and this enabled Alison to identify 710 unique protein clusters in S. tubifer that may be relevant to symbiosis. Interestingly, functional enrichment analysis of these clusters using Gene Ontology (GO) categories highlights that S. tubifer is enriched in genes involved in host-microbe interactions and visceral muscle development.

From these findings, Alison questions whether bioluminescent cardinalfish have evolved changes in musculature that contribute to the development of the gut-associated light organ. To further identify genes relevant to symbiosis, Alison wishes to use RNAseq to transcriptionally profile the S. tubifer light organ, and to use gene-editing technology to delete candidate genes and therefore empirically test their role in symbiosis.
A preprint of the chromosomal-level genome assembly of Siphamia tubifer is available on bioRxiv.
The African BioGenome Project
One of the most highly anticipated sessions at Biodiversity Genomics 2021 was the African BioGenome Project, which is a pan-African collaboration, which brings together scientists from all over the African continent “to address challenges to food systems and help protect African biodiversity through genomics”. ThankGod Ebeneezer of EMBL-EBI presented the African BioGenome Project and DAISEA (Digital Information in Africa for a Sustainable Agri-Environment) Grand Challenges, which include the following:
- Genomic and bioinformatics technologies for agri-environment
- Crops and livestock improvement and health
- Genomics for conservation of endangered and endemic species
- Socioeconomic and policy
- Technology development, knowledge exchange and industry
Simplice Nouala, who is Head of the Agricultural and Food Security Division at the African Union Commission, highlighted the need for Africa-wide continental support to genotype the resources that the African continent has. As Simplice explained, “The real challenge that we are having today is to know what we have. We know we have but we do not know what we have”. As Simplice puts it, from an African perspective the strategic objective is “to map and conserve our main resources.”
We look forward to hearing more from this amazing initiative which aims to improve and sustain biodiversity across Africa, and helping publish and share many more genomes coming from across the globe and taxonomic tree. For more on our genomics publishing workflow in GigaByte see our talk from last years meeting.