Cell Biology in Washington DC: GigaScience at ASCB/EMBO Cell Bio 2022

The Washington DC Walter E. Washington Convention Center was the venue for the recent Cell Bio 2022 Meeting that took place on 3-7 December 2022. Cell Bio 2022, the joint meeting of the American Society for Cell Biology (ASCB) and European Molecular Biology Organization (EMBO), is the largest yearly gathering for the Cell Biology community. GigaScience Data Scientist Chris Armit is a regular attendee of this event (see 2019 and 2020 write-ups) and reports on the impact of Cell Biology research in expanding our understanding of living systems.
The Challenge of Proteomics in Living Cells
In a Keynote talk entitled “Chemogenetic and optogenetic technologies for probing molecular and cellular networks”, Alice Ting (Stanford University) highlighted the use of proximity labelling for the study of endogenous proteins and RNA in living cells with high spatial and temporal accuracy. A focus of this work is the study of endogenous untagged proteins that is scalable to entire proteomes. Towards this end, Alice and colleagues have developed various “spray painting” methods that utilise enzymes to tag proteins with biotin labels in the neighbourhood of specific organelles.
Proximity labeling was followed by mass spectrometry to identify the biotinylated tagged proteins. Alice was swift to point out that the reducing environment of the cytosol did represent a challenge for proximate-dependent tagging. For example, the reducing environment is problematic for horseradish peroxidase, which is a frequently used enzyme for fixed samples.
The enzyme ascorbate peroxidase is far superior for living cells, but as with horseradish peroxidase this enzyme still requires hydrogen peroxide as a substrate, which is notoriously toxic for cells. Alice and colleagues are currently exploring the utility of laccase, which is an enzyme that uses oxygen rather than hydrogen peroxide as a substrate, and is potentially non-toxic. As Alice explains in her Keynote talk, one of the key advantages of laccase is that “it has the potential to be much more in vivo compatible”.
Mini-guts as a model of Symmetry Breaking
EMBO Gold Medal 2022 winner Prisca Liberali (Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland) gave a talk entitled “Decoding the design principles of tissue organisation”. Prisca explained that tissues are not constructed machines, but are built up from short and long range interactions. Using intestinal organoids as a model system – or as Prisca refers to them ‘mini-guts’ – an imaging approach was used for feature extraction and the ability to explore the changing geometry of specific enterocyte cells in each and every organoid.
The derivation of organoid lines is, in itself, intriguing. Single cell isolation using FACS is used to isolate Lgr5 positive cells. Lgr5 is a known stem cell marker of the intestine, and the Lgr5 positive cell is the ancestor of all cell types in the intestine. Lgr5 positive cells are then seeded to matrigel cultures in 96-well plates, and the enterocysts that develop share some of the hallmarks of the intestine in vivo, including a constellation of different cell types with distinct geometry.
Mini-gut self-organisation of a stem cell niche
Interestingly, Lgr5 expression is lost early during organoid development, but returns as the organoid enterocyst begins to differentiate. This is interpreted as the cultured mini-guts activating a regenerative response. The mechanism of symmetry breaking – whereby distinct cell types differentiate from a uniform population of cells – is partially understood, and it is known that the Hippo pathway activates Yap1 as a sensor of cellular states. Specifically, Yap1 triggers activation of Notch/Delta lateral inhibition, with the Yap1 positive cell also expressing Dll1 (Delta).
Symmetry breaking occurs at the 16-32 cell stage, at which point Lgr5 positive cells are also appearing in the organoid enterocyst. Consequently, the creation of the Lgr5 positive stem cell niche is a regenerative response that can be explored in intestinal organoid culture. This technology may offer invaluable insights for understanding tissue injury and repair.
For more on Organoids and Organogenesis, see our recent Blog of VIZBI 2022.
Making waves: Scaling up hexagonal arrays in Zebrafish
Continuing the focus on the importance of short and long range interactions, Maya Evanitsky (Di Talia lab, Duke University) gave an interesting talk on zebrafish scale development. Zebrafish scales are made of calcified matrix and are highly organised in a hexagonal array. So how can cellular signalling accomplish such a feat? There are certain mutant strains of zebrafish – such as the Ectodysplasin mutant – that have no scales, and this is a clue to understanding how scale formation is coordinated.
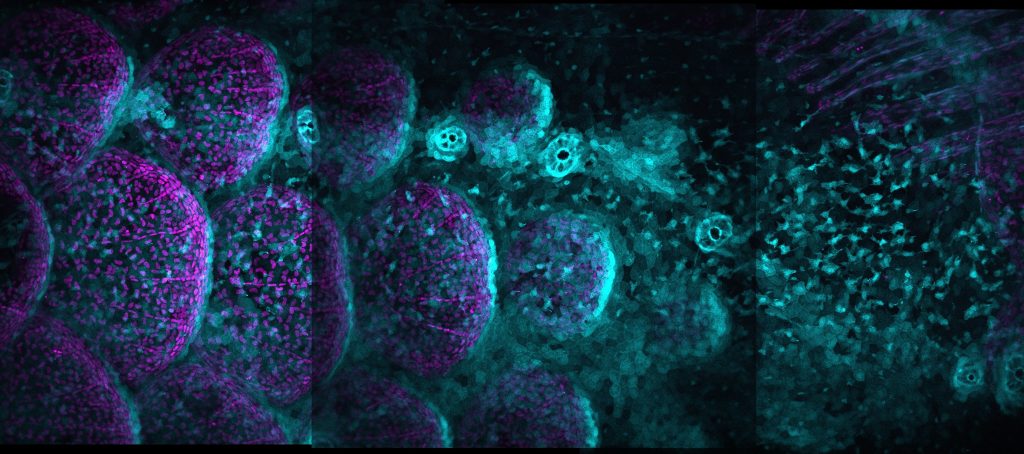
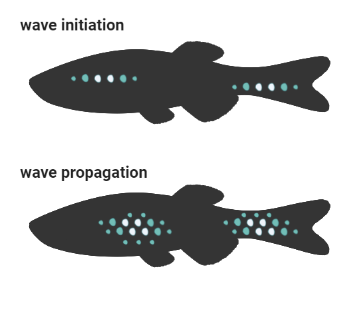
Ectodysplasin protein signals through the NF-κB pathway and induces nuclear translocation of the transcription factor NF-κB. Through use of videomicroscopy, Maya showed that NF-κB signalling is propagated as a travelling wave through a primordial population of osteoblast-like scale-forming cells.
Interestingly, cuts in scale tissue block propagation of this wave, and create a delay in scale growth. As Maya explains, through wave initiation and wave propagation, NF-κB signalling regulates scale patterning through a positive feedback loop.
For more on zebrafish embryogenesis, see Life Unfolding inside the Cell in our recent Blog of VIZBI 2022.
Nature’s Toughest Bug: Radioresistance in the Tardigrade
Ionising radiation is directly genotoxic and causes single strand and double strand DNA breaks. In addition, ionising radiation generates reactive oxygen species that create apurinic / apyrimidinic sites in DNA that require Base Excision Repair (BER).
The tardigrade is an eight-legged, segmented micro-animal that has the extraordinary ability to withstand what for other animals are lethal doses of ionising radiation. Courtney Clark-Hachtel (UNC Chapel Hill) gave a fascinating talk on the secrets of radiation resistance in this tiny creature, and highlighted that a 100 Gray dose of ionising radiation does not affect reproduction and survival in nature’s toughest bug.
Attribution: Goldstein lab – tardigrades, CC BY-SA 2.0, via Wikimedia Commons
Through use of TUNEL labelling, which is more commonly used as a method to detect double strand breaks in apoptotic cells, Courtney was able to demonstrate that tardigrades do show extensive DNA damage in the form of double strand DNA breaks immediately following ionising radiation insult, but that these are repaired swiftly.
Through use of RNA-seq, Courtney further demonstrated that the tardigrade showed a robust transcriptional stress response with an incredible >250-fold upregulation in DNA repair pathway genes following genotoxic insult. To provide some necessary context, in mammals a 4-8-fold upregulation in DNA repair pathway genes is what a researcher would normally observe.
The critical role of DNA repair in surviving radiation damage
To further investigate the role of individual DNA repair pathway genes, Courtney used RNAi to knockdown candidate genes, and to explore the effect on survival to ionising radiation insult. In this way, Courtney was able to demonstrate that the double strand DNA repair pathway gene Ku80 was critical for the survival of tardigrades in response to radiation damage.
What I find especially puzzling about the tardigrade is its very short lifespan. Courtney highlighted that these micro-animals only live to about 3 months, and I do wonder whether there is an evolutionary trade-off between a super-charged DNA damage response and longevity. Whether there are longer-term detrimental effects to physiology in radiation-resistant micro-animals is a question that remains unanswered.
For more on Evolutionary Genomics, see our Blog on Biodiversity Genomics 2021.
Combating Neurodegenerative Disease and Genome instability in Cancer
“…it turns out that cells of the nervous system hadn’t read the textbooks” – Don Cleveland
Don Cleveland (Ludwig Institute, University of California, San Diego) delivered the E.B. Wilson Medal Lecture entitled “From Tau to evolution of genome instability in cancer”. It is the 45th anniversary of the discovery of Tau, which causes neurofibrillary tangles associated with Alzheimer’s Disease. Misfolded Tau protein is also observed in Chronic Traumatic Encephalopathy, which is linked to repeated head injuries and is an additional cause of dementia.
Don highlighted the controversy surrounding early attempts to deliver DNA into the nervous system as treatments for neurodegenerative disease. The consensus at the time was that this was impossible, and Don quips that “it turns out that cells of the nervous system hadn’t read the textbooks”.
In a pioneering study, Don and colleagues used gene silencing as a treatment for Amyotrophic Lateral Sclerosis (ALS), a neurodegenerative disease that leads to loss of muscle control. The strategy was to use catalytic mRNA degradation and altered RNA splicing polyadenylation to regulate transcriptional levels of disease-associated genes. Don highlighted that his treatment had 3-4 months efficacy.
Similar gene silencing approaches were used for Spinal Muscular Atrophy through targeting SMN2 splicing, and Parkinson’s Disease via lowering of levels of SNCA, which encodes the presynaptic neuronal protein α-Synuclein linked to Parkinson’s disease.
Understanding aneuploidy
On the subject of genome instability, Don additionally focused on chromothripsis, which is the shattering of chromosomes leading to aneuploidy and large-scale chromosomal rearrangement.
In experimental chromothripsis, centromere inactivation induces micronucleation and chromosome shattering, and this leads to intra- and inter-chromosomal rearrangements but also circularised extrachromosomal DNA (ecDNA). I was intrigued to find out that chromothripsis drives ecDNA formation in methotrexate resistance, and that the ecDNA evolves through continuing chromothripsis. This means that, over time, ecDNA becomes massively rearranged.
In an effort to suppress gene amplification – a phenomenon often observed in human cancer – Don questions whether we can target chromothripsis and ecDNA evolution. Towards this end, Don highlighted the dominant role of the endonuclease N4BP2 in chromothripsis. N4BP2 has 5′-polynucleotide kinase and nicking endonuclease activity, and induces DNA damage in ruptured micronuclei. In addition, the absence of N4BP2 strongly inhibits fragmentation of a micronucleated Y chromosome. This is an excellent candidate gene for augmenting genome instability in cancer.
Open Data and Tools in Education
At GigaScience Press we see immense value in Citizen Science (see recent Blog on the fight against mosquito borne disease), and the “Open Data and Tools in Education” Workshop as presented by the Allen Institute was of immense interest. As panelist Kaitlyn Casimo, who leads the Allen Institute education program explained, the Allen Institute for Brain Science is “developing a parts list” of cell types in the brain. Panelist Gideon Dunster additionally highlighted that a focus of the Allen Institute for Cell Science is “recognising patterns in large-scale complex data”.
As an important Cell Biology resource, the Allen Institute has made a library of 3D images of 200,000 cells publicly available. In addition, educators have used Allen Institute online image analysis tools to ask questions such as ‘What happens when cell division goes wrong?’ by inviting school students to add and subtract cell structures to study their relationships during specific cell divisions.
Gaming interface for Citizen Scientists
For budding Neuroscientists, Mozak Brainbuillder uses a gaming interface to allow school students to annotate very large neuronal images. This tool can be used in the classroom to help understand neuron diversity ‘beyond the textbook’ through generation of 3D reconstructions from open-source data.
Of key importance here is familiarising school-level students with real-life data. Illinois high school teacher Tom Martinez – who was one of the panelists – highlighted how these online tools are of immense benefit to his students through “correcting misconceptions with cell-specific imagery”. As Tom further explained, “the imagery is different from what they’re used to from textbooks”.
Tom has the impressive career achievement of successfully campaigning for a cell culture lab in his school, where students from the age of 13 are educated in cell culture technology. Tom’s is the only school in Illinois with this facility, and this is a pioneering approach for inspiring future Cell Biologists.
For more on Citizen Science, see our GigaBlog series. GigaScience is also highlighting studies taking imaging into the 3rd Dimension
ASCB/EMBO Cell Bio 2023 is scheduled for December 2-6, 2023, in Boston, MA.