Improvements for Man and Machine in Scientific Publishing
Frictionless Data improves not just machine readability of scientific articles, but also enables humans to directly interact with the data within the article itself. A new article in GigaByte demonstrates a new workflow where frictionless data can help bring papers to life with interactive figures.
The need for information from research outputs to be more findable, accessible, interoperable, and reusable (FAIR) has spurred researchers, database managers, and publishers to continually look for new and better ways to make information machine-readable. Another equally important area is creating articles that readers can actively engage with, rather than passively taking in information from reading a published article. One tool that easily improves machine readability of data is a data standard called Frictionless Data, developed by the Open Knowledge Foundation. Published today in GigaByte a new Technical Release paper demonstrates that not only does Frictionless Data drastically improve machine readability, it can also turn normally static figures within the article into dynamic entities that allow readers to directly interact with the data within the article. Demonstrating that the use of Frictionless Data can tackle two important activities: allowing both man and machine to use and directly engage with scientific outputs in a dynamic fashion.
Integration of Frictionless Data was carried out on an article by a team of researchers from the University of Melbourne led by Professor Anthony Papenfuss, whose lab have been long time advocates of open and reproducible research (being the first authors with GigaScience to integrate protocols.io protocols into their papers back in 2016). Making sure the data, source code, and every other sharable component of their research is openly available to the community. This makes their work especially amenable to utilising new tools on top of their articles to make the published work dynamic and actively usable. The article here presents two new open source tools, svaRetro and svaNUMT, for interpreting difficult to structural variation in genome analysis. These help annotate novel genomic events that are missed in most genome assembly pipelines: such as retrotransposition events and insertion of DNA fragments from the mitochondria to the nuclear DNA, which contribute to the complexity of genome sequences and the understanding of gene function and genome evolution.
Commenting on why they follow these open practices the authors say: “Reproducibility is a cornerstone of science and a particular focus for us and our research. We want to make our data and analyses reproducible and open so that they are accessible by others, and easy to use and build upon. We hope that others find our methods useful, and that our analyses and data are reused in future developments such as benchmarking studies.”
Frictionless Data smoothening the flow of Open Science
The openness and availability of all of the research components behind these tools and analyses created a perfect opportunity to implement Frictionless Data to make the article far more machine readable. During the process of adding this to the article, Raniere Silva from City University of Hong Kong, as part of a FAIR data internship, made the fortuitous discovery that Frictionless Data could also play a role in improving human interaction with the article. You can get a behind-the-scenes and more detailed look of his work this summer in a guest blog. The figures, for the first time, were regenerated in an interactive manner. In the example here, readers can not only view the summary information presented in the figure, they can hover over data points to see the exact numbers and information behind these, and also manipulate the figure itself to view specific components that are of interest.
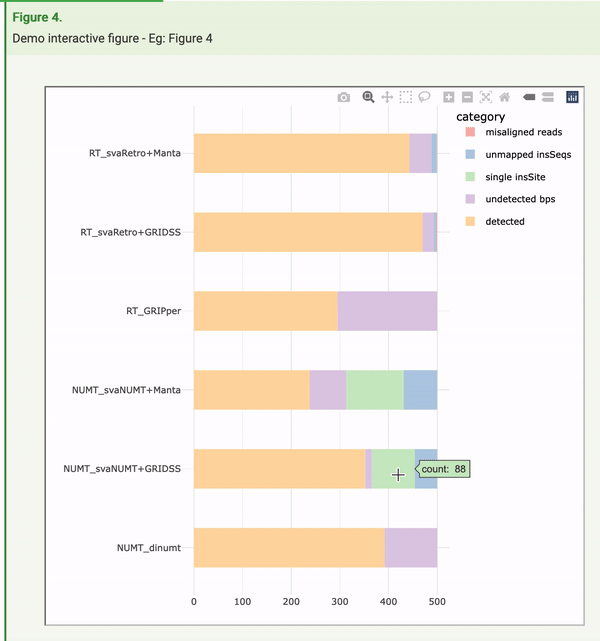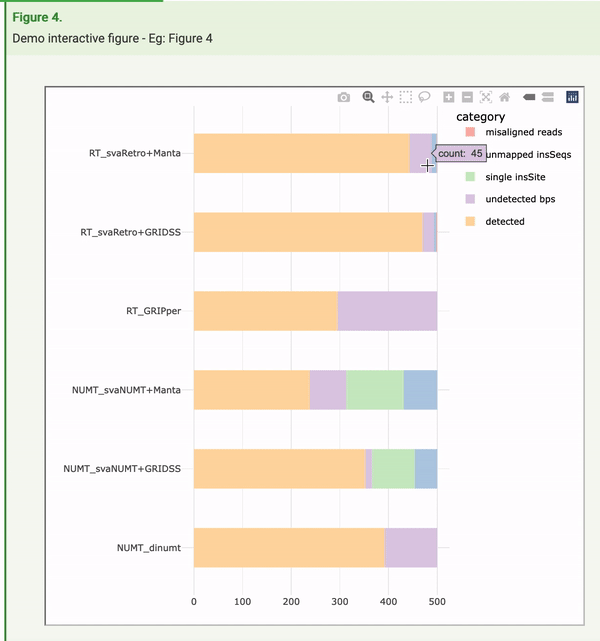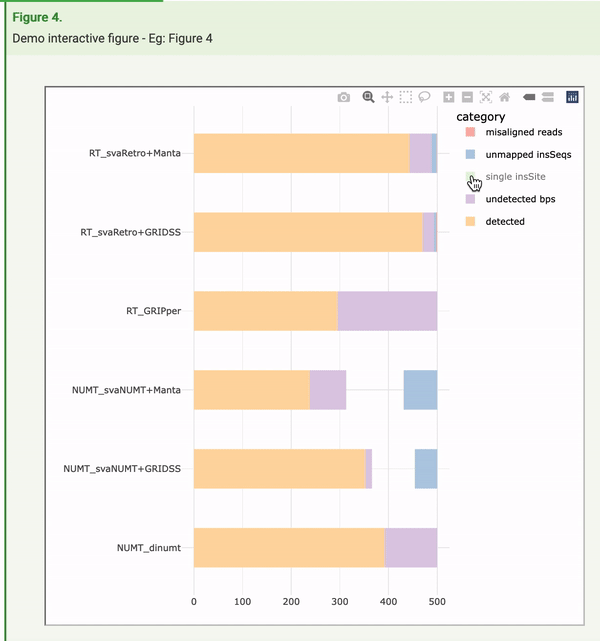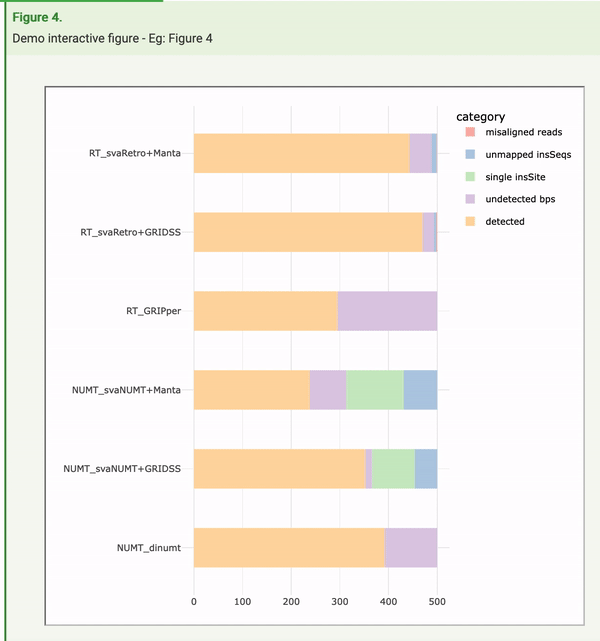
Ranier says: “My biggest surprise was that the Frictionless Data Package specifications in conjunction with the popular Plotly tool has functions to convert a static visualisation into a dynamic one. This massively reduces the barrier for many researchers to produce dynamic data visualisation as they only need to add a line or two to their code. GigaByte made a huge leap by publishing the dynamic data visualisation and I hope it inspires other journals to publish dynamic data visualisation.”
When asked what they found most useful from this process, the authors stated: “The interactive figures are a great addition to the paper. We found the interactive functions made reading labels easier, especially for label-rich figures, and liked that the figures were accessible in SVG format, allowing viewing and editing without losing information from the figures.”
To promote the use of Frictionless Data in more published articles, Raniere wrote a detailed handbook that includes an introduction to the use of Frictionless Data, an introduction to the specifications, short working examples for creating an author’s own data package, and long examples, based on published articles in GigaScience and GigaByte journals, illustrating the creation and use of Frictionless Data. The goal is for the handbook to serve as the start of a conversation within the scientific community of how to embrace Frictionless Data. This handbook also provides a resource and guidance to make things easier and for data producers to submit articles with these packages to data publishers, such as GigaScience Press.
Of added interest, in addition to the inclusion of Frictionless Data, paper is that for the first time as the figures were regenerated in an interactive manner this process combined a CODECHECK certificate of reproducible computation. We’ve previously written about CODECHECK and showcased a few examples of these certificates in GigaScience, but this is the first time the CODECHECKing process has been worked into the generation of interactive figures. And also the first example used in the review and publication process of a GigaByte paper.

And I, for one, welcome our future machine readers
The use of Frictionless Data and all the downstream elements it enables, serves as transformative steps in scientific publishing, as they improve machine readability and reproducibility, and turn scientific articles from their old-fashioned static format into a 21st century living document. Increasing trust and interaction for our human readers through interactive figures has long been a goal of GigaScience Press (see our 2014 attempts at “push button” papers using Knitr and our showcasing last year of Stencila’s Executable Research Article format). This new approach demonstrates an integrated way of achieving this in our GigaByte papers using the novel XML-first publishing workflow from River Valley Technologies. With the Frictionless Data package file for our machine readers to better interact with and understand the data and software available in our GigaDB repository. These types of novel, data-literate additions to the publication process are part of the reason GigaByte was the winner of the 2022 ALSPS Innovation in Publishing Award last month.
To encourage more authors to use Frictionless Data in their articles, all manuscripts submitted to GigaByte before the end of 2022 that include Frictionless Data examples will be given a free APC (currently $350). Authors interested in finding out how to do this should contact the editors at GigaByte at editorial@gigabytejournal.com.
Further Reading:
Dong R, Cameron D, Bedo J, Papenfuss AT. (2022). svaRetro and svaNUMT: modular packages for annotating retrotransposed transcripts and nuclear integration of mitochondrial DNA in genome sequencing data. GigaByte. 2022. https://doi.org/10.46471/gigabyte.70
Dong R; Cameron D; Papenfuss AT (2022): Supporting data for “svaRetro and svaNUMT: modular packages for annotating retrotransposed transcripts and nuclear integration of mitochondrial DNA in genome sequencing data” GigaScience Database. http://dx.doi.org/10.5524/102318
Raniere Gaia Costa da Silva. (2022). CODECHECK Certificate 2022-018. Zenodo. https://doi.org/10.5281/zenodo.7084333