Tag Archives: microbial genomics
Open CycloneSeq Benchmarking For Complete Bacterial Genomes
Scott Edmunds - April 25, 2025
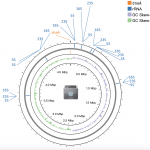
As another DNA Day treat GigaByte today publishes new benchmark data and analysis of the new CycloneSEQ platform, a novel sequencing technology using nanopores that demonstrates here the ability to sequence complete bacterial genomes. Following on from the recent official launch of BGI’s new CycloneSEQ single-molecule sequencer the new Data Release published today in GigaByte […]
One group of bacteria to rule them all
Hans Zauner - March 26, 2021
New research in GigaScience reveals a previously hidden diversity of symbiotic Rickettsia bacteria. The new data are also relevant for studying human and animal diseases that are caused by some types of Rickettsia.
Genomics Standards on the Danube. GigaScience at GSC21 in Vienna
Chris Armit - June 3, 2019

The 21st meeting of the Genomics Standards Consortium (GSC21) took place last week in Vienna at one of the oldest universities in the world – the University of Vienna – from May 20th-23rd. We’ve been long time supporters and participants of the Genomics Standards Consortium meetings going back to 2012’s GSC13 in Shenzhen, and have […]
Mock the Metagenome. Author Q&A with Nick Loman & Sam Nicholls
Scott Edmunds - May 15, 2019

The mock metagenome, MAGs and breaking the first rule of Long Read Club Out today in GigaScience is a new “mock metagenome” Data Note from the Nick Loman lab in Birmingham showcasing the latest long-read sequencing technologies from Oxford Nanopore. Having published the first nanopore E. coli genome with us in 2014 showcasing the then […]
Reprocessing the Microbial Genomic Goldmine: Winner of the ICG13 Prize
Scott Edmunds - December 13, 2018

Out today is the winner of our ICG13 Prize, presenting work that can aid in revealing new biologically relevant findings and missed genes from previously generated transcriptome assemblies. Teaching old data new tricks, and maximising every last nugget of information from previously funded research. Here we present some insight into why the reviewers and judges […]