The (first) GigaScience Press End-of-Year Review
This end-of-the-year festive summary is different from previous ones: For the first time, we can look back at a full publication year of our new baby, GigaByte journal. The older sibling GigaScience will also get the attention it deserves, before celebrating its 10th birthday in 2022.
Let’s first have a look at how the new family member is doing.

The excellent news is that GigaByte is getting attention from authors. The journal published 35 articles so far, with supporting open data and code in our database GigaDB and signed reviews openly available. The concept of GigaByte clearly meets a need: to rapidly and easily share accounts of research outputs (including large data and open code), allowing others to build upon the results even before there’s a complete “story” to tell.
Another piece of cheerful GigaByte-related news is that we were able to keep the promise of extremely rapid publication after editorial acceptance. Thanks to the end-to-end pipeline provided by our partner RiverValley we usually can publish articles within days after editorial acceptance. And for reviewers, we make their task easier with a questionnaire-based assessment, also designed to speed up the process.
In terms of topics, GigaByte proved attractive to scientists from diverse fields, with genomics being particularly popular. Examples of papers published in 2021 include genome resources for the critically endangered woylie, the North American water vole, the humpback puffer fish, the mule deer and many other species. On the software side, GigaByte’s Technical Release format is also becoming popular, as evidenced by a paper on SMARTdenovo, a single-molecule sequencing assembler.

But the scope of the journal is much broader than genomics, and GigaByte for example also published X-ray micro-tomographic data of live larvae of a beetle – and we’d love to see more imaging-related submissions in the future.
Call for papers: Biodiversity of human disease vectors.
Looking ahead to the coming months, biodiversity datasets will be a priority for GigaByte, including geolocation information linked to other sample-based data such as DNA sequences or morphological data. We are excited that the Global Biodiversity Information Facility (GBIF) decided to launch a special issue, publishing new datasets presenting biodiversity data for research on vectors of human diseases. The project is supported by TDR, the Special Programme for Research and Training in Tropical Diseases, hosted at the World Health Organization.

As we recently wrote in our announcement of the collaboration in GigaBlog, “GigaByte’s novel, end-to-end XML publishing platform, means publication can be done in a quicker and more cost-effective manner better designed for these more granular research objects that don’t require such a labour intensive and detailed vehicle for sharing. It also allows additional interactivity and we can work with the authors to embed maps, video and imaging data plugins and other relevant tools for visualizing data and results in the final publications.”
If you are working on biodiversity of vectors of human disease we hope you will be interested to submit to this special issue. Please read the call for papers here.
Play with code within a research paper
GigaByte and RiverValley’s flexible publishing pipeline are a great sandbox for showcasing new features that go beyond the classical research article – such as in the form of Executable Research Papers. Using technology from Stencila and Code Ocean enables readers to use two different platforms to inspect the code, to modify it, and then to re-execute it directly within the article, which thus becomes a living document (see a first example here).
In the spirit of moving away from the traditional Research Article as a static object, we also consider Updates to previous work and the first GigaByte publication of this type was published in June. As our Editor-in-Chief explained in a blog post, Update Articles allow authors to truly “publish at the speed of research”, as GigaByte’s motto goes:
“ As a way to publish new data or software outputs from a previously published article at a time point where there is useful information to share but there are little-to-no new analyses, conceptual findings, or major software upgrades.”
Gigantum: Transparency and Reproducibility at large scale
Integrating useful external tools into journal articles was always one of our favorite things, as evidenced by our previous cross-links with the reproducibility platform Code Ocean and the Galaxy project. This year, Gigantum joined the reproducibility toolkit, both for GigaScience and GigaByte. Gigantum is an open source web application that aims for better collaboration and sharing of work that is rich in data and code. We are seeing more and more complex submissions from the Machine Learning world, supported by big, diverse datasets that come alongside custom-made code in need of powerful computational infrastructure. Gigantum is a great way to improve reproducibility and transparency for this type of computationally intensive work, and the first authors who tested the incorporation of the platform with GigaScience were excited about the possibilities it has to offer.
Here’s how it works:
Cite the code, cite the paper
Also in the spirit of moving beyond traditional research articles and giving credit to other research outputs, such as code and data, we followed with interest the new software citation guidelines of the Joint Declaration of Data Citation Principles that came out of FORCE11 Community meetings and workshops. We already supported citation of software and code of course, but the new guidelines bring some additional improvements, such as including the software version in the citation, as well as a recommendation that both, the software repository and the article describing it (if available) should be cited.
Open Access and Peer Review weeks
This year’s Open Access Week (Oct 23-31) had the theme “ It Matters How We Open Knowledge: Building Structural Equity”, and we published a post looking back over 10 of our favorite GigaScience papers providing examples of barriers we’ve tried to break for more open science.
September’s Peer Review week covered the theme “Identity in Peer Review”, and we posted on the OUP blog and contributed a video to the Peer Review Week Youtube channel our experiences on this topic from carrying out open, named peer review.
Using this as an opportunity to announce that GigaScience and GigaByte have implemented the “Transparent Review in Peer-review” or TRiP workflow that enables journals to post peer reviews alongside the preprints of submitted manuscripts (see our blog for more).
For more on the topic see the Open Publishing Fest webinar we participated in covering preprint peer review.
Virtual meetings and cake
COVID still has the world in its grip (some parts more than others) and most conferences were held in virtual form, again – including ISMB, where we traditionally celebrate GigaScience’s birthday. Alas, it was another digital birthday party, featuring a video to celebrate 9 years of GigaScience, and a virtual birthday cake-cutting by our publisher Laurie Goodman.
With so many meetings being held virtually it meant the team could widen their reach and participate and present at many of them organised across the globe. Our Editor in Chief Scott presented a keynote at LanBix (Sri Lankan bioinformatics) conference on software publishing and citation in the COVID-19 era (see the video here). As well as webinars from EBSCO on Research Intelligence: Open Research, Open Data and Open Publishing (watch here) and COPE (the Committee on Publication Ethics) on Ethical practice in research data publication (watch here). We also just presented a community talk and poster at FORCE2021 on the interactivity of our GigaByte papers.
One of the few meetings that did take place in-person was the 7th annual Women in Science Conference in Qingdao, a satellite meeting of BGI’s International Conference on Genomics (ICG). Again organized by our Publishing Director Laurie Goodman, and co-chaired by Ying Gu of BGI Research, the meeting brought together both young and more experienced female researchers to present their work and to discuss the deeper issues that affect women in science.
Our data scientists Chris Armit attended a couple of meetings to keep us up-tp-date on new developments in areas such as Data Management (NASEM), Biodiversity genomics, Neuroinformatics (INCF) and Biological Visualization (VIZBI).
Of ducks and snails. Some 2021 research highlights from GigaScience
No summary of the year would be complete without celebrating some of the amazing science we published in 2021. GigaScience continues to be popular with authors who wish to share their accounts of exceptional, high quality genome assemblies (including supporting data in GigaDB) and this year we again have some truly special genomes: For example, the mediterranean cone snail genome that Rafael Zardoya and colleagues published.
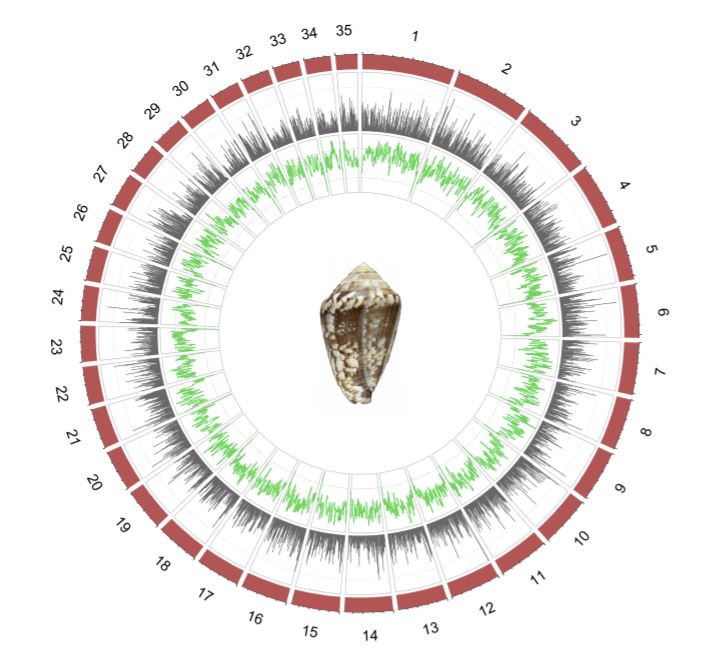
It’s one of the largest gastropod genomes sequenced so far, with 3.59 billion base pairs organized in 35 chromosomes. Cone snails are infamous for their diverse toxins, and the new genome assembly allowed the authors to explore the evolutionary origins of cone snail venom, as Zardoya explained for GigaBlog: “For many years, it was puzzling how the great diversity of cone snail toxins was generated. The genome answers this question: Many gene copies distributed throughout the different chromosomes are producing this diversity, which is later enhanced by post-translational modifications of the peptides. “
Pekin duck
New milestones were also achieved in bird genomics: Spread out over several journals, a number of articles appeared presenting a new generation of high-quality avian genomes at chromosome level – with Pekin duck as one of these new high-end bird assemblies being published in GigaScience.
The Peking duck genome will help to study the domestication of poultry, and it’s an also important model organism investigating virus transmission and control.
Genetic diversity in Ukraine
In May, Taras Oleksyk and colleagues from Ukraine, the US and China published the largest study of human genetic diversity in Ukraine. Ukraine is made up of a population formed via millennia of migration, and the territory served as a key prehistoric and historic crossroads for the spread of humans across Europe and into Asia. Dr Oleksyk:
“Ukraine accounts for roughly a quarter of the genetic variation documented in Europe. It’s a part of the world that cannot be ignored in future genetic and biomedical studies.”
AI for mental health assessment
Machine learning work is becoming a cornerstone of GigaScience’s content (see the section on Gigantum above) and ML methods are changing basically all of science – including psychology. In a recent GigaScience article, Denis Engemann and his team demonstrated that machine learning can yield “proxy measures” for brain-related health issues from large population data, without the need for a specialist’s assessment (Paper here, author Q&A here). Using questionnaire and imaging data available from the UK biobank was crucial for this project, which was covered for a wider audience in the French weekly magazine “L’Express.”
Open imaging data to assist AI in the COVID-19 fight
The National COVID-19 Chest Imaging Database (NCCID) is a centralized database containing chest X-rays, Computed Tomography (CT) and MRI scans from patients across the UK. Announced in a GigaScienne data note in November, the database is “one of the largest datasets of its kind and it is continuously growing, with over 27 contributing hospitals and imaging data from over 10,000 patients in the training set”, as the author explained for GigaBlog recently.

The data will be particularly useful in the fight against COVID; which brings us finally to the outlook to the New Year 2022 – and the hope that the pandemic will be better under control, despite some variant-related worries hanging in the sky.
One thing we are looking forward to next summer is GigaScience’s 10th birthday, and if the pandemic allows, we very much would like to eat real cake with real people to celebrate the occasion. Watch out for a special issue of GigaScience (also available in print), looking back at 10 successful years and looking forward to the future of open access publishing.
Almost 10 years of GigaScience also signifies 10 years of great support from reviewers and editorial board members. A big thank you for your interest, your help and advice in 2021, and I hope you continue to work with us in 2022 and beyond.
We wish you and all our authors and readers a Happy New Year!