Harvesting the final fruits of the plant tree of life
 Lessons learned in the evolution of large-scale data sharing
Lessons learned in the evolution of large-scale data sharing
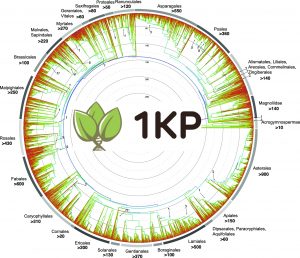
New studies out today elucidate the framework for 1 billion years of green plant evolution. The work are the results of nearly a decades work from an international consortium of nearly 200 plant researchers generating gene sequences from more than 1100 plant species. Here and in an accompanying Data Note on the 1KP capstone data we provide a behind-the-scenes look at how this was carried out.
The 1KP: One Thousand Plant Transcriptomes Initiative (or more accurately the 1.2KP) is a mammoth effort to examine the diversification of plant species, genes and genomes across the more than one-billion-year history of green plants dating back to the ancestors of flowering plants and green algae. In 2014 the initial pilot based on the phylogenomics analysis of the first 85 species was released, and accompanying this we published a useful guide for the data access for the ongoing project. Recent decades have seen the rise of these “mega-sequencing” projects, and in 2009 the Toronto Data Release Workshop affirmed the commitments to prepublication release of large genomic data sets which were originally developed in Human Genome Project. The resulting Toronto Statement encouraged data producers to produce a citable statement in which they describe the data set and their intentions in respect of analysis and publication. The original GigaScience paper filled that requirement, linking to the treasure trove of raw and processed data that supported the project that have been building in the EBI, Cyverse, and our GigaDB repository.
Five years on its nice to see this project on to its conclusion with the capstone analysis in Nature, and an accompanying GigaScience Data Note again describes the whole data set and provides additional details on the sample and sequence processing as well as quality assessments of the 1KP capstone data. Adding value by providing a user guide for data re-use, links to detailed protocols (thanks to protocols.io for their help in inputting the 18 different RNA-extraction methods in), and a lot of other behind-the-scenes information. like the contamination analysis. These details are useful for the box-set completists and data re-users, rather than the casual readers of the capstone paper that are only interested in the narrative.
This narrative fascinating though in throwing light on the complicated and billion year journey of the evolution of plants. The findings in Nature, reveal the complex timing of whole genome duplications and the origins, expansions and contractions of gene families contributing to fundamental genetic innovations enabling the evolution of green algae, mosses, ferns, conifer trees, flowering plants and all other green plant lineages. The history of how and when plants secured the ability to grow tall, and make seeds, flowers and fruits provides a framework for understanding plant diversity around the planet including annual crops and long-lived forest tree species.
The study inspired a community effort to gather and sequence diverse plant lineages derived from terrestrial and aquatic habitats on a global scale. Open to everyone, over 100 taxonomic specialists contributed material from field and living collections that include the Central Collection of Algal Cultures, Royal Botanic Gardens, Kew, Royal Botanic Garden Edinburgh, Atlanta Botanical Garden, New York Botanical Garden, Fairylake Botanical Garden, Shenzhen, The Florida Museum of Natural History, Duke University, University of British Columbia Botanical Garden and The University of Alberta. Size and scale or participation and sampling matters. By sequencing and analyzing genes from a broad sampling of plant species, researchers are better able to reconstruct gene content in the ancestors of all crops and model plant species, and gain a more complete picture of the gene and genome duplications that enabled evolutionary innovations. A decade ago such an ambitious and multi-participatory project on 1,000 species was very new, and other than individual genome projects there was little in the way of role models on how to carry this out. On top of proving a great example of the way to carry out open and collaborative large-scale research, it also provided an interesting model of how such work could be supported outside of traditional funding streams that were starting to lose interest in (non-clinical) genome projects.
Project instigator Gane Ka-Shu Wong organized private funding through the Somekh Family Foundation as well as support from the Government of Alberta and a sequencing commitment from BGI to launch 1KP. Once the project was operational, additional computational and data storage resources came from other ongoing projects, including CyVerse (formerly iPlant). The massive scope of the project demanded development and refinement of completely new computational tools for sequence assembly and phylogenetic analysis, including the SOAPdenovo-trans de novo transcriptome assembler designed specifically for the project that has subsequently been a extremely popular tool for many in the RNA-seq community.

Gane Ka-Shu Wong presenting 1KP & 10KP results at the PAG Asia workshop we participated in.
“New algorithms were developed by software engineers at BGI to assemble the massive volume of gene sequence data generated for this project,” explained Wong. With the computational outputs of much of this currently being written and reviewed as Data Notes in a similar manner to the phylogenomic resources we published alongside the Avian Phylogenomic Project, that also aimed to resolve the phylogeny of major branch of the tree of life (see GigaBlog). Some of these are already coming out as pre-prints, for example this work in bioRxiv covering the whole genome duplications.
Also involved in the Avian Phylogenomic Project, Computer scientists Tandy Warnow of the University of Illinois and Siavash Mirarab of the University of California San Diego developed new algorithms for inferring evolutionary relationships from hundreds of gene sequences for over one thousand species, addressing substantial heterogeneity in evolutionary histories across the genomes.
“Gene family expansions through duplication events catalyzed diversification of plant form and function across the green tree of life,” said co-author Marcel Quint, professor of crop physiology, at Halle University, Germany. “Such expansions unleashed during terrestrialization or even before set the stage for evolutionary innovations including the origin of the seed and later the origin of the flower.”
“The view of evolutionary relationships provided by 1KP has led to new hypotheses about the origins of key structures and processes in green plants,” said co-author Pam Soltis, of the Florida Museum of Natural History, University of Florida.
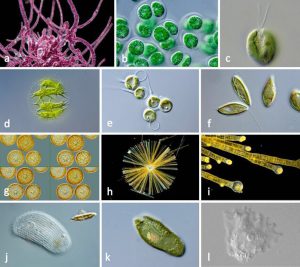
Understanding the huge diversity of protists is one of the focuses of the follow on 10KP project. Source 1KP doi:10.1093/gigascience/giy013
From 1K to 10K (and beyond)
With the 1KP reaching its conclusion, many of the team are now scaling up to whole genomes and larger sample sizes with the 10KP project that we published an announcement paper on last year. This aims to sequence complete genomes from more than 10,000 plants and protists, and address fundamental questions in plant evolution and diversity. Building an annotated reference genome for a member of every genus of the land plants and green algae, as well as a phylodiverse set of species representing both photosynthetic and heterotrophic protists.
With even bigger challenges regarding taxonomic sampling and collection for 10KP, a pilot of this was carried out to provide insight into the feasibility and technical requirements for such “planetary-scale” genome projects. This was the digitization of Ruili botanical garden genome project that we published earlier this year, and presented on with many of the 1KP and 10KP consortium in a workshop at the PAG Asia meeting in June (see above picture and GigaBlog). Watch this space on the continuing legacy of this work, and see how the offspring of 1KP and other large scale phylogenomics projects covering other corners of the tree of life continues to bear fruit. For more on this project see the 1KP website (www.onekp.com), where information on the continued outputs is being catalogued and collected.
References
Carpenter et al. Access to RNA-sequencing data from 1,173 plant species: The 1000 Plant transcriptomes initiative (1KP). Gigascience. 2019. doi:10.1093/gigascience/giz126
One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature, 574:2019, doi: 10.1038/s41586-019-1693-2
Cheng S, Melkonian M, Smith SA, Brockington S, Archibald JM, Delaux PM, Li FW, Melkonian B, Mavrodiev EV, Sun W, Fu Y, Yang H, Soltis DE, Graham SW, Soltis PS, Liu X, Xu X, Wong GK. 10KP: A phylodiverse genome sequencing plan. Gigascience. 2018 Mar 1;7(3):1-9. doi: 10.1093/gigascience/giy013.