A Passion for Morphing Passiflora Leaves: Author Q&A with Daniel Chitwood
 Passiflora, commonly known as Passion Vines or Passion Flowers is a genus encompassing around 500 species, all of which exhibit such huge variation in leaf shape. To further understand the unique diversity of Passiflora leaves, a recent paper published in GigaScience, presents a morphometric analysis and unique open dataset encompassing over 3,300 leaves from 40 different Passiflora species. The research was carried out by two researchers; Daniel Chitwood, formerly of the Donald Danforth Plant Science Center, and Wagner Otoni from Universidade Federal de Vicosa, Brasil. Here we present a Q&A with lead author, Dan, who shares his thoughts on the importance of Passiflora and morphometric analyses in plant phenomics.
Passiflora, commonly known as Passion Vines or Passion Flowers is a genus encompassing around 500 species, all of which exhibit such huge variation in leaf shape. To further understand the unique diversity of Passiflora leaves, a recent paper published in GigaScience, presents a morphometric analysis and unique open dataset encompassing over 3,300 leaves from 40 different Passiflora species. The research was carried out by two researchers; Daniel Chitwood, formerly of the Donald Danforth Plant Science Center, and Wagner Otoni from Universidade Federal de Vicosa, Brasil. Here we present a Q&A with lead author, Dan, who shares his thoughts on the importance of Passiflora and morphometric analyses in plant phenomics.
Why is Passiflora of interest to you?
Passiflora species exhibit impressive variation in leaf shape, which makes them an ideal group to test and compare morphometric techniques with. Beyond diverse shapes, Passiflora species are also vines, which makes keeping track of the nodes the leaves arise from a simple task. Some Passiflora vines dramatically change leaf shape within an individual plant and the juvenile and adult leaves may have very different shapes. Because leaves vary so dramatically between species and within vines, Passiflora species are the perfect model for describing the genetic and developmental basis of leaf shape.
You mention the diversity in Passiflora leaves in your manuscript and it is this reason why you have chosen to do morphometric analysis on their leaves – why do you think there is so much diversity in Passiflora leaf shape?
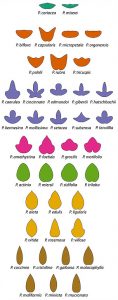
Fig 3. Chitwood and Otoni, GigaScience (2017) 6 (1): 1-13
Although leaf shape has a strong genetic, developmental, and environmental basis, the underlying evolutionary pressures leading to diverse shapes are often difficult to uncover. Passiflora leaves are unique in that Heliconius butterflies lay their eggs on them. One defense of Passiflora species is mimicry, and sometimes leaves possess “false eggs”, small yellow structures that deter butterflies from laying their eggs. Some have speculated that the diverse leaf shapes among Passiflora species are to evade the butterflies, as work has shown that the butterflies can remember leaf shape. The sometimes dramatic changes in leaf shape within a vine may serve a similar purpose. Although the causes of Passiflora leaf shape ultimately remain unknown, there are many interesting hypotheses!
The dataset supporting this paper is 14.31 GB in size; what were the challenges with handling such a large dataset of images?
The raw morphometric data, landmark coordinates and the harmonic series from Elliptical Fourier Descriptors, that serves as the basis for all statistical analyses, is actually very small. However, it is critical that the original, high resolution scans are accessible to others. 14.31 GB of scans is too large to include in any traditional supplemental data, and repositories for high resolution scans, such as these, are either too specialized to take leaf images or will not accept such a large amount of data (without a fee). Further, it is easier to coordinate the availability of data with manuscript submission at the same journal. Accessibility of the data to others is the main challenge working with a large dataset such as this.
From a broader plant phenomics point of view; how do you envision people reusing such a unique dataset?
The interpretation of plant phenomic data remains problematic. Morphometrics is a time-honored technique to analyze complex phenomic data. There are many different techniques to compare, some of which may be more appropriate than others to analyze data. By making the raw images available, it is our hope that different groups can apply disparate morphometric techniques to the exact same shapes so that an empirical comparison of different approaches can be made.
Dan’s research is published as part of our Plant Phenomics series (Guest Edited by Rubén Rellán Álvarez, Guillaume Lobet, Malia Gehan, and Srikant Srinivasan), which aims to shed light on new advances, applications, and challenges, and to improve data sharing, integration, analyses and reproducibility in plant phenomics. Another paper recently published in this series, is Phenobook – an open source tool that can help researchers in different field locations to organize, collect, and save experimental data for further analyses.
 Although improvements in plant phenomics research has created many new opportunities to understand and predict dynamic phenotypic responses to genotypic variation. However, problems and opportunities have emerged, such as the ability to collect phenotypic data in a way that can be combined with different datasets for meta-analyses, as well as the generation of models or as ground-truth datasets. And with thanks to the 3,000 Rice genomes and 183 millet genomes serving as published examples of genotype data – there is now a strong need for high-throughput plant phenotyping data in these specific research communities.
Although improvements in plant phenomics research has created many new opportunities to understand and predict dynamic phenotypic responses to genotypic variation. However, problems and opportunities have emerged, such as the ability to collect phenotypic data in a way that can be combined with different datasets for meta-analyses, as well as the generation of models or as ground-truth datasets. And with thanks to the 3,000 Rice genomes and 183 millet genomes serving as published examples of genotype data – there is now a strong need for high-throughput plant phenotyping data in these specific research communities.
We encourage the submission of Research Articles and Technical Notes, as well as Data Notes, which are papers that focus on the description of interesting plant phenomic datasets, curated and hosted in our database, GigaDB. We also consider Commentaries and thought provoking Reviews in this area.
Potential topics include, but are not limited to:
- New methods in phenotype data collection e.g. Drones, imaging techniques
- New tools in phenotype data integration and analyses
- Morphometrics
- Organ-scale phenomics
- Databases, management and workflows
- Research or Data wrapped in a containerized form e.g. Docker, BioBox, VMs
Submit your manuscript, or email editorial@gigasciencejournal.com for more information.