Democratising Data: The African Orphan Crops Consortium & International Data Week
Pressing Challenges for the Global Research Community
Continuous growth of the world population is expected to double the worldwide demand for food by 2050. Eighty-eight percent of countries currently face a serious burden of malnutrition, especially in Africa and South-East Asia. To diversify and stabilize global food supply, enhance agricultural productivity and tackle malnutrition, greater use of neglected or underutilized local plants through democratising data and research on them is required. Researchers on the ground need to be involved in this, but in a survey of academics and students in sub-Saharan Africa, 80% polled reported that they have held unpaid research positions. Open Access is essential for them, but how are Article Processing Charge models going to work in a world where journals can charge over $5,000 US dollars?

Monkey business at the IDW coffee breaks.
Attempting to answer this question, GigaScience organised a session at the International Data Week conference in Gaborone, Botswana last month. Bringing together the Research Data Alliance and CODATA meetings that we have previously attended, hosting the meeting in Africa for the first time was a brilliant opportunity to bring together different global perspectives and approaches to research data together. The conference was opened by the Botswanan President Dr Mokgweetsi Masisi, and delegates were extremely impressed to see a head of state talking about the importances of research data management. With Over 850 attendees coming from 66 countries, and 60% African participation, for it was refreshing to reach a critical mass of diverse voices that were not the “usual subjects” from the well funded European and North American institutions, and everyone who made it to Botswana felt it was a historic and eye opening meeting because of it. Other than vervet monkeys raiding the coffee breaks (pictured), the conference followed its usual structure and themes and was of extremely high quality.

Chris Armit discussing the democratising of data.
While these meetings usually have a generic data publishing track we wanted to use the novel location and audience to do something a little different, specifically organising a track on “Democratising Data Publishing: A Global Perspective”. With GigaScience Data Scientist Chris Armit starting the track off and providing some scene setting (see his slides here), we then heard Chinese perspectives to Data Publishing from Jianhui Li (China Scientific Data and ScienceDB) and the Nature Publishing view from Varsha Khodiyar (Scientific Data). We then heard from more infrastructure related perspectives, our Editorial Board Member Susanna Sansone talked about FAIRsharing and how making digital objects FAIR can democratise data publishing (see slides), Mark Parsons on Power and Persistent Identifiers, and Daniel Mietchen on how distributed Wikidata and Wikibase approaches could tackle the democratizing issue (slides here). We ended with a more local perspective, Anwar Vahed giving a very interesting overview on the South African national research data infrastructure: DIRISA. To give some time to discuss the topic in hand, reiterating the challenges, but more importantly propose any more democratic ways ahead beyond the APC, we ended with a panel. This featured several of the speakers and added Elizabeth Marincola and Rebecca Lawrence to give us some insight into how the African Academy of Sciences Open Research journal and platform is trying to tackle this.
Rice fills the agricultural data gap
The specific example that Chris focussed on was agricultural data and the Rice3K project we published the first outputs of data in 2014. With the aim of improving the productivity of the the crop that provides the majority of daily calories for much of the worlds poor. The 13.4TB of data quadrupled the amount of rice genomic data in the public domain, but this and the resulting 100TB of processed data made available in AWS, while open was impractical to download and use by most researchers due to computer bandwidth and cost. What was data publishing did help democratise was the early release of data, incentivised by the Data Note article in GigaScience that helped the data producers to get early publication credit 4 years before the analysis paper was published (which eventually appeared in Nature this year).
While downloading and using compute on the raw and partially processed data was prohibitively expensive for those working on rice breeding in the regions dependent on the crop, researchers at IRRI have now managed to genotype and analyse it down to such a level that it can be cost effectively shared. The RiceGalaxy platform also has a GUI to allow plant breeders to utilise this genetic data without coding skills, and is currently running via AWS Singapore & local servers at a cost of about $100 USD/month. This CGIAR Excellence in Plant Breeding model is now rolling out to other crops, demonstrating there is a way that the benefits of these big-data approaches can indeed be democratised. This work was one of the six winners of our ICG-prize track in October, and you can read the preprint and watch the video of Venice Juanillas presenting on our GigaTV youtube channel.
The African Orphan Crops Consortium bears its first fruits
Following the example set by these rice genomes, other agricultural crop species with great potential for genome-assisted improvement are being sequenced by a number of consortia. The goal of the African Orphan Crops Consortium (AOCC), an international public–private partnership, primarily comprises African Union’s New Partnership for Africa’s Development (AU-NEPAD Agency); World Agroforestry Centre (ICRAF); BGI; and University of California, Davis. With generous support from its partners, AOCC is working on sequencing the genomes of 101 plants that contribute to traditional African food supplies by 2020. These neglected or orphan plants have been seldom studied by scientists, but are of major importance in many African countries. They are usually grown by smallholder farmers, either for consumption or local sale, and are a major food source for 600 million rural Africans.
Our co-publishers BGI has completed reference genome analysis for six crops, five of which are published in a new Data Note in GigaScience today. With six genomes near to completion, and nearly 30 more in progress, expect to see more fruits of this work being released in the near future. The first five plants include the legume crops the Hyacinth Bean (Lablab purpureus), the Bambara Groundnut (Vigna subterranea), and larger tree species such as the Apple-Ring Acacia (Faidherbia albida), Marula (Sclerocarya birrea) and the medicinal Horseradish Tree (Moringa oleifera). All of which are important for food, fodder, shelter and medicinal properties for smallholder farmers across Africa. Unlike many agricultural staple species that have complex hybrid genomes, the assembled genome sizes were relatively compact: 535 Mb, 395Mb, 653Mb, 330Mb, 216Mb respectively, accounting for 97.3%, 93.5%, 98.9%, 92.9%, 77.9% of the estimated genome size respectively.
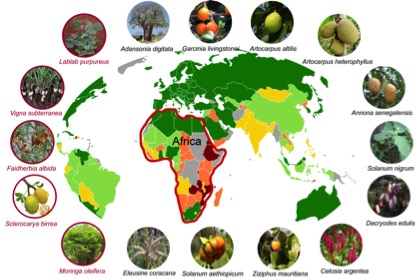
This comprehensive study in GigaScience reports the sequencing, assembly, and annotation of five genomes of underutilized plants in Africa, along with details of their key evolutionary features. Following the example set by rice, the draft genomes of these species will serve as an important complementary resource for non-model food crops, especially the leguminous plants, and will be valuable for both agroforestry and evolutionary research. Improving these underutilized plants using genomics-assisted tools and methods could help to bring food security to millions of people. Author and AOCC consortium member Prasad Hendre was supposed to present in our International Data Week Democratising Data session, but as he unfortunately couldn’t make his talk some of this work was also previewed in Chris Armit’s presentation. Meeting the goals of democratisation outlined above, as the research is published Open Access it can be read by researchers across the world without restriction, and all the data is released without restriction for plant researchers and breeders via NCBI and GigaDB. The protocols are available in protocols.io and data of these first five species is available for download from the via the following DOIs:
Genomic data of the Apple-Ring Acacia (Faidherbia albida). http://dx.doi.org/10.5524/101054
Genomic data of the Bambara Groundnut (Vigna subterranea). http://dx.doi.org/10.5524/101055
Genomic data of the Hyacinth Bean (Lablab purpureus). http://dx.doi.org/10.5524/101056
Genomic data of Marula (Sclerocarya birrea). http://dx.doi.org/10.5524/101057
Genomic data of the Horseradish Tree (Moringa oleifera). http://dx.doi.org/10.5524/101058
Further Reading:
Chang Y et al. The draft genomes of five agriculturally important African orphan crops. GigaScience. 2018. doi: 10.1093/gigascience/giy152
3,000 rice genomes project. The 3,000 rice genomes project. Gigascience. 2014 May 28;3:7. doi: 10.1186/2047-217X-3-7