PAG Asia 2019: Plant and Animal Genomes (with Chinese Characteristics)
Regular readers will know GigaScience has published a lot of plant and animal genomes, and the biggest conference for this research community is the appropriately named Plant & Animal Genome Conference (PAG). We’ve attended a number of these giant meetings at their San Diego base, and in recent years they have been branching out to host more convenient satellites in Asia (PAG Asia). Initially organised in Singapore, last month we went along to PAG Asia as the first time its been hosted on our doorstep in China. Not as big as the main meeting, PAG Asia 2019 still had an impressive 354 attendees from 24 different countries. On top of attending in the Futian Shangri-La, we also participated in one of the workshops, and here is our write-up of how it went.
At #PAGAsia19 in Shenzhen for the rest of this week. BGI’s Xu Xun is a co-chair of the meeting and introducing keynote Ning Yang pic.twitter.com/EG8l2TEt0j
— GigaScience (@GigaScience) June 6, 2019
DNA Zoo and the $1000 Genome Assembly
With one million species at risk of extinction the is an urgent need to capture as much genomic data as we can about these disappearing species to monitor populations and inform conservation, captive breeding and re-wilding strategies. Mammalian species are particularly at risk, with 25% endangered or threatened. This was the rationale of DNA Zoo workshop that opened the meeting in Shenzhen. While GigaScience has worked with and published many of the outputs of the Genome10K (10,000 vertebrate genomes) consortium that is now focusing on “platinum-level” chromosome-scale genomes and constantly testing and throwing together the latest state-of-the-art genomics technologies, DNA Zoo are aiming at a slightly different approach. Focusing on a single cost effective technology (Hi-C) to maximize what they can achieve in the current limited funding environment. Our recent African painted-dog genome paper that utilized 10X data got close to their $1,000 ball park de novo genome (costing roughly $3,000 USD), and their technological approach will hopefully bring these costs down further while still providing high-quality reference genomes. Following our approach at released bird genomes via social media, the DNA Zoo team have been releasing their nearly 100 genomic datasets via their blog.
Erez saying they’ve already shared 78 mammalian chromosome scale assemblies on https://t.co/dmYTZe5EA9. Releasing one genome a week, all released without restriction #PAGAsia19 pic.twitter.com/MUEPp0nLd2
— GigaScience (@GigaScience) June 6, 2019
Erez Liberman Aiden of Baylor College of Medicine – who developed the Hi-C chromosome conformation capture technique – delivered the opening talk for this session and explained how Hi-C, which was primarily developed for understanding 3D genome architecture, has become a novel, low-cost solution for delivering a chromosomal level genome assembly. Hi-C allows locations that were approximate in 3D space to become adjacent through crosslinking of chromatin. Crosslinked chromatin is digested into fragments using restriction enzymes, and the crosslinked fragments are then ligated together. By sequencing these fragments, researchers obtain a map that helps us understand chromosomal folding in the nucleus. As Erez points out, the overwhelming finding from these Hi-C maps is that chromosomes tend to fold on themselves. Indeed, as Erez explained, “pairs of loci on the same chromosome tend to bump into each other at a high rate”. DNA Zoo have utilised this phenomenon to develop low-cost chromosomal level genome assemblies. This hybrid assembly approach uses a combination of Illumina short-read sequences, which are insufficient for chromosome-level assemblies, combined with Hi-C data of the same species. Using this approach, the 3D genome maps inform the sequence assembly at a fraction of the price compared to using long-read sequencing and/or optical map technology. Olga Dudchenko – also from Baylor College of Medicine – further explained that it was possible to upgrade a $1K end-to-end assembly with, for example, PacBio long reads to produce “platinum assemblies”. Olga additionally presented an overview of the automated pipeline software that DNA Zoo use to assemble genomes. Parwinder Kaur of University of Western Australia further explained how these low-cost technologies will increase our understanding of genetic biodiversity in Australia, which includes an astonishing 87% of mammals endemic to the continent. We are big fans of “community genomes”, and to get the ball rolling sequencing some of their amazing endemic wildlife Parwinder has recruited Chokka the Quokka to crowdfund one of Australia’s most “selfie-genic” animals.
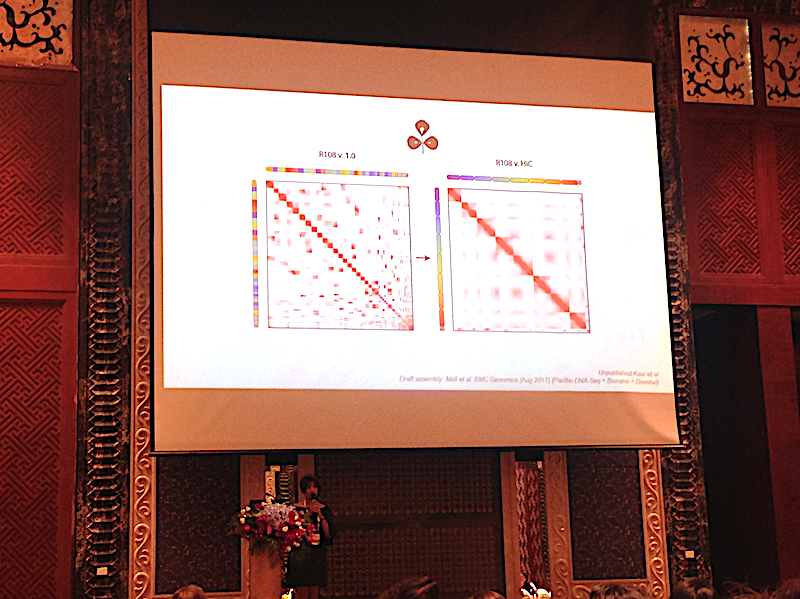
Parwinder Kaur of University of Western Australia explains how Hi-C is enabling low-cost chromosomal genome assembly, and how this further impacts on our understanding of genetic biodiversity in Australia.
Thinking big with 10KP and the Ruili Botanical Garden
Gane Ka-Shu-Wong of University of Alberta and BGI-Shenzhen, and Xin Liu of BGI-Shenzhen introduced the 10,000 Plant Genomes Project (10KP, of which we published the announcement paper), which aims to sequence a vast amount of plant genomes representing every major clade of plants and eukaryotic microbes. Gane explained that from a genomics perspective there is a focus on long-range contiguity, and further highlighted that novel genes found in land plants are also found in algae, which may suggest that terrestrialization could have begun with the algae. Gane also reported that preliminary findings from 1KP (1,000 Plants Project, of which we published the data access paper) suggest an unexpected abundance of horizontal gene transfer from soil bacteria to algae. Michael Melkonian of the University of Cologne offered deeper insights into algae – and suggested tripartite endosymbiois between cyanobacteria, Chlamydia sp., and a eukaryotic species as one possibility for algae’s evolutionary origins – and was quick to point out that these often overlooked single-cell plants are responsible for 50% of global photosynthesis. Yang Liu of BGI-Shezhen reported on mosses as a potential source of evolutionary novelties such as the stomata and cuticle, which are essential for drought resistance. Yang Liu additionally highlighted the impressive cold and heat resistance traits that can be found in the Bryophyta that allows them to survive in glaciers and geothermal areas.
Great to see @Bauhinia genome discussed at #PAGAsia19, Stephen Tsui presenting in the 10KP track on using their data to assemble Bauhinia genomes. You can see the slides here https://t.co/foPaScN2eY pic.twitter.com/dbBEBjD5f3
— BauhiniaGenome (@BauhiniaGenome) June 8, 2019
Huan Liu (BGI-Shenzhen), Stephen Kwok-Wing Tsui (Chinese University of Hong Kong) and GigaScience Executive Editor Scott Edmunds further reported on the hugely impressive (molecular and imaging) digitization of the 500-hectare Botanical Garden in Ruili in subtropical Yunnan province in China. You may have read the recently published research, and this talk provided behind the scenes insight into how this mammoth project came about. Scott explains, the 689 vascular plant species sequenced “effectively triples the number of plant species with available genome data”. The first part of the project from Huan Liu explained how this provided a nice to investing the feasibility and technical requirements for “planetary-scale” projects such as the 10KP. Scott then covered the data management and organisation to make this data discoverable and re-usable. The final part was of a downstream user perspective, Stephen Tsui showing how he could re-use this unassembled data for the community Hong Kong Bauhinia Genome project we have been working with him on. For more insight you can see the slides here.

Scott Edmunds of GigaScience explains how the Ruili Botanical Garden dataset was captured.
Crop Informatics
Ken McNally of IRRI (International Rice Research Institute) introduced a diverse selection of talks that reported on the current use of informatics in rice genomics. Star Yanxin Gao of Cornell University highlighted the Genomic Open-Source Breeding Informatics Tool Portal that couples genomic information with phenomic data to enhance decision making in the breeding of staple crops. In addition, Shuhui Song of the Beijing Institute of Genomics introduced the BIG Data Center’s Rice Variation Database and highlighted its ability to assess reannotation results as part of its role in the Information Commons for Rice (IC4R). Following this, Dmytro Chebotarov of IRRI reported on SNP-Seek, which is a web application that enables efficient querying of large rice genomic datasets, such as the 3,000 Rice Genomes (Rice3K) data that we previously published in a Data Note. SNP-Seek incorporates analysis and visualization tools that include a genome browser with SNP, gene and structural variants tracks, phylogenetics tree and MDS plot visualizations. The Rice Galaxy server presenting this data being one of the winners of our ICG Prize last year and presented by first author Venice Juanillas in our session at the conference (see video below). The democratising potential of this approach was demonstrated previously by the GigaScience team at International Data Week in Gaborone, Botswana and it was equally well received at PAG Asia 2019.
PAG Asia 2020 is tentatively scheduled for Shenzhen again, and if we do not see some of you there we will be at the main PAG in San Diego in January 2020 and likely provide coverage of how these and other plant and animal genome projects are proceeding.