Plant and Animal Genomes (PAG) 31st annual conference
PAG (Plant and Animal Genomes conference) returned to the Town and Country resort San Diego for its 31st installment this January (Jan 12-17, 2024), bigger and better than ever before! The GigaScience Press team are regular attendees of the meeting (see last years write-up), and this year members of our Editorial and Curation teams joined nearly 3000 delegates from over 60 countries.
With a leisurely start to the conference on the Friday around 10am, my first stop was to reacquaint myself with the fine work of the Oxford Nanopore Technology (ONT) team at their ORG.one session. Kara Dicks introduced the ORG.one project, which is an ONT funded project to assist with at-location sequencing (of plant and animal genomes) of threatened or endangered species. They will provide the tools free of charge to enable the generation of approximately 20x genome coverage of any species listed on the IUCN red list as endangered (status EN or CR).
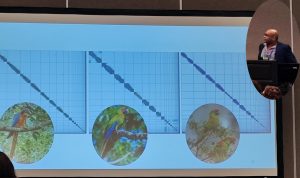
Three parrot genom HiC assembly plots
Then followed a series of talks on some of the species already sequenced through this project including the; Butternut (an endangered species of Walnut in USA); the Widemouth gambusia (a sulphide adapted species of fish in Mexico); the Eastern Hoolock gibbon of China; as well as what may well have been the most entertaining presentation by final year PhD student Taylor Hains on 6 genomes of endangered Macaws and Cockatoos from across the world (especially as GigaScience has a soft spot for parrot genomes). Taylor presented with great passion for the subject showing in equal measure both excitement and despair! Apparently, avian genomes differ from mammalian genomes in that they commonly contain many many micro-chromosomes (or to use Taylors wording “Parrots have messed up chromosomes!”) that are almost impossible to identify by karyotyping. This difference can cause issues (often resulting in tears) with the informatics tools which are generally designed for use with mammal genomes.
Whether the micro-chromosomes are the reason or not has yet to be determined, but a feature of parrots is that they are able to mate across species boundaries, and regularly do (again in Taylors inimitable style “Biology doesn’t apply to parrots they are just horny little bastards!”. This is a feature that results in a multitude of different hybrids which tend to be very popular with the pet trade as they can be highly unusual and beautiful specimens.
Data reuse
Data reuse was a recurrent theme in many of the sessions. GigaScience Editorial Board Member (EBM) Mike Schatz’spresentation included a mention of the GTEx project, which is now 10 years old, and is essentially a reanalysis of 311TB of raw data hosted in DBGap, which has been accessed by >250 groups around the world. Some simple maths shows that if all 250 groups had downloaded the raw data to re-do the analysis themselves it would have meant >100PB of data had been stored in multiple duplicates. This is simply not a scaleable solution, hence the need to run the analysis in the data storage environment rather than copy large data files to a place to run the analysis. AnVIL is one such tool that enables multiple groups to work on 1 copy of data in the cloud.
The AgBioData consortium have a Data reuse working group, which presented on the importance of data quality, incomplete metadata, and lack of incentives for researchers to provide it. They advocate for a common standard across all genomes to be adopted to enable easier sharing and comparison. They also highlighted a paper published in 2021 (sorry I missed the reference) that showed >50% of cases where the author had indicated that data is available on request but had failed to provide those data when requested.
Plenaries
Despite the fact the conference started on Friday morning, we had to wait until Sunday evening for the first plenary speaker, Appolinaire Djikeng, who preceded the opening reception in the exhibition hall. Among many accolades mentioned in his introduction, Appolinaire was a recipient of the Nelson Mandela Justice award in 2020 for his work in global agricultural development. The topic of his presentation at PAG was “Livestock and the Food Systems: A Focus on Smallholder Systems in the Global South”. Appolinaire outlined various challenges that we still face in the global food system particularly related to food security, as well as highlighting the recent advances that are helping meet those challenges. A key feature of the talk was the different approaches to farming in different countries, particularly the tendency for the Global North to create large fields with little or no crop diversity compared to the smallholder style farming predominant in the Global South. And we should be aware that the right solution to these challenges may be different in those different settings.
Over the following 3 days we received 6 more plenary talks from distinguished scientists from around the world. Lucy Van Dorp presented virtually on the subject of tracking pathogens in space and time, using the recent COVID-19 pandemic as an example of data-rich reconstruction of the evolution of the virus genome through time.
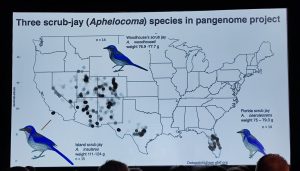
Distribution of three scrub-jay species in USA
Scott Edwards said “More people know me from my cycling across the USA than for my science!” with reference to his epic 2020 bicycle ride from coast to coast in aid of Black Lives Matter. In fact he is a well-renowned curator at the Museum of Comparative Zoology at Harvard with a passion for ornithology. Scott gave an overview of the current state of affairs in population pangenomics in birds, including the scrub-jay pangenome project (pictured).
Machine Learning
In line with the current trends in all areas of science, there were a good deal of talks using or discussing the potential of machine learning technologies. For example, Chris Mungall spoke about “Using Large Language Models to Build Ontologies”. Due to a delayed flight, Chris actually arrived at the meeting room during the question time of the speaker preceding him, (some may say perfect timing) but that didn’t fluster him in the slightest. With hundreds of ontologies in use in plant and animal genomics it’s a challenge to keep them up-to-date, so can LLMS (large language models) help with that? In short, Chris seems to think there is a great potential for it to, yes, particularly if Retrivall Augmented Generation (RAG) tools can be used to get more accurate results e.g. DRAGON AI. Experiments were performed to check how well the DRAGON AI-generated term definitions compared to the human-written ones by blind review and ranking. The result was that the human-generated definitions were on average more understandable and accurate than the AI-generated ones. However, there is scope to use AI to fill the gaps with some human oversight and checks. Chris stressed that these tools will not replace humans but could lead to greater productivity.
In a similar vein, Aleksey Zimin presented “Data Beats Machine Learning for Genome Annotation”. Zimin’s group examined the use of machine learning techniques to annotate genomes and found that they do provide an increased coverage, but generally at a cost to accuracy particularly in the UTRs. He proposes a tool called “eviAnn” which uses protein evidence from related species to annotate genomes.
Citizen Science
As followers of GigaScience Press will know, we are great advocates of citizen science projects, so I was delighted to attend the “Participatory and Citizen Science Genomics” session. For me the highlight of the session was “The INCREASE Citizen Science Project (www.pulsesincrease.eu)”. Kerstin Neumann presented an overview of the project which in summary aims to revive (genetic) diversity from gene banks back into the community for the common bean. It is now entering its 4th year with a steady increase in participation from across Europe. It’s a mixture of social science (community building) and phenomics. Citizen participants are each provided with a selection of 5 different heritage seed lines from a seed bank along with 1 control line (the same line is given to every participant) and asked to grow them at home and record various traits in an app. The community participants are encouraged to discuss challenges with one another via a Facebook group which reduces the administrative burden of dealing with direct queries. The community aims to foster seed sharing from person to person. Each participant is expected to save some seed from each harvest to regrow and/or share with others, while any excess produce is for their personal consumption. The app includes 36 traits split by the level of experience of the users from beginners to experts, as this is a Europe-wide project, it’s also translated into various European languages to enable communication with participants. At present this project is only available within Europe due to the issues with import/export of living materials.
The end
For many of us, the science ended rather abruptly when the fire alarm at the conference centre went off during the penultimate talk of the last session just before lunch. Speaking to Helen Brabham after her talk (which was one of those interrupted by the fire alarm) it appears most participants (including myself I must admit! Sorry Helen) opted to get lunch on the lawn after the fire alarm instead of heading back into the session. Despite this slightly disappointing end to the working day, it was not all over yet! As tradition states, the final night of PAG is the conference banquet, where the exhibition hall is turned into a giant banqueting hall replete with dance floor and DJ where everyone makes the most of the last chance to network.
Thanks to the organisers for providing the Press pass, and I look forward to seeing everyone again at PAG32!
For those that cannot wait an entire year, there are now alternative PAG conferences around the world, PAG-Asia and PAG-Australia will be held in 2024, with plans for PAG-India being made for 2025.