Cracking sex chromosome evolution via egg-laying birds and mammals

Today is a big day for the genomics of egg-laying animals, with the coordinated publication of three papers on animal sex chromosome evolution and diversity in GigaScience, Nature, and Genome Research by the Qi Zhou lab at Zhejiang University. Also co-lead by Frank Grutzner from Adelaide and GigaScience Editorial Board Member Guojie Zhang of Copenhagen University, this collection of studies includes two chromosome-level monotreme genomes (platypus and echidna) in Nature, and two chromosome scale bird genomes for the Pekin Duck in GigaScience and Emu in Genome Research. These three works systematically depict the dazzling diversity of animal sex chromosomes, and use Hi-C technology to gain new insight into the origin and evolution of sex chromosomes at the level of the three-dimensional genome.
New advances in genomics finally fit the bill
Monotremes are unusual species which display a unique mix of mammalian and reptilian features and form the most distantly related group of living mammals. While the some of these species may have a superficial resemblance in the way they look and lay eggs, it is a fun coincidence that the duck and duck-billed platypus both join the chromosome-scale genome club on the same day. This work discovering other characteristics of birds, with monotremes found to express one of the three vitellogenin genes which encode the major egg proteins responsible for making the yolk in chicken, while these genes have become lost in other mammals. On top of these fascinating physical traits, monotremes and birds are equally interesting models of sex-chromosome evolution, and this has been the overlapping focus of these three new papers. Bringing together the recent huge improvements in sequencing with new computational methods specifically developed for this work, this has allowed the researchers to obtain the highest quality chromosome-level genome assemblies that now enable the resolution of these previously too challenging-to-assemble sex chromosomes.
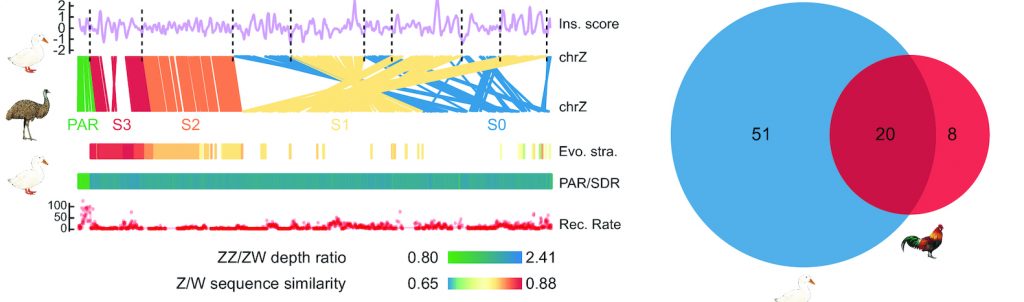
Monotremes have a complex sex chromosome system consisting of five pairs of sex chromosomes, and their X chromosomes share no homology with human’s X-chromosome, having more in common with the avian Z-chromosome (see more in this interview with Qi Zhou talking about the Nature paper). Alongside characterizing these in monotremes, for the first time the Z and W sex chromosomes have also been assembled in the duck, discovering they have more than twice the number of W-chromosome genes than the chicken. As with monotremes, bird genomes can have some very interesting characteristics from an evolutionary perspective, with some of them even demonstrating fusions between autosomal and sex chromosome elements. In 2019 we published an example of this mechanism of sex-determination and speciation, via a paper that discovered novel neo-sex chromosomes in the eastern yellow robin (see GigaBlog).
Looking deeper at the work in our new GigaScience paper, Pekin duck on top of being one of the most commercially important poultry species, has a higher tolerance to avian influenza than chicken. Making it also an important model organism for the study of virus transmission and control. From the perspective of evolutionary biology, Pekin duck, chicken, goose, quail and other poultry form the Gallonanserae Superorder, which is one of the three major branches of modern birds. Obtaining a high-quality duck genome at chromosome level therefore is of great significance for studying the domestication of poultry and filling an important gap in resolving phylogeny of vertebrates and birds in projects such as the VGP (Vertebrate Genome Project) and B10K (Bird 10,000 Genomes project, as we’ve covered here before).

Duck genome tales
In 2013 researchers at our co-publishers BGI published the first draft of the Pekin duck genome. However, due to the lack of genetic map and usage of short reads, most of the genome sequences and genes in the draft, especially the highly repetitive sex chromosome sequence, were in a state of fragmentation, Meaning the exact chromosomal position was not clear. This new update uses third generation pacbio long reading sequencing technology, combined with the state-of-the-art 10x, Bionano and Hi-C chromatin conformation capture technology to assemble and annotate the new reference genome, and located more than 95% of the sequences to specific chromosome positions. The new genome (named ZJU1.0) has achieved a 60-fold increase in N50 continuity (76.3MB vs. 1.2MB) compared with the previously published version released in GigaDB (BGI1.0), and also corrected the incomplete and incorrect annotations caused by repetitive sequences.
Avian genomes soar into the third dimension
The new Pekin duck and emu genomes are among the first of a new generation of high-quality avian genomes assembled at chromosome level. With assembly quality of these genomes equal to or better than that of the most studied avian models of chicken and zebrafinch. Furthermore, through the comparative analysis of 3D genome data, the researchers divided the 3D spatially associated domain (TAD) of Pekin duck, and found that most of the regulatory domains were highly conserved between chicken and duck, reflecting the strong selection effect of natural selection on regulatory domains. Although there were a lot of rearrangements between chicken and duck genomes, these rearrangements tended not to destroy the internal structure of these regulatory domains.
The evolution of duck sex
Importantly this work marks the first publication of the Z and W sex chromosome sequences of the duck. Previous studies have shown that genes related to sex traits, such as egg laying traits, tend to be enriched on sex chromosomes. Therefore, these newly published sequences will provide important references for the domestication of sex related traits in Pekin duck. In addition, the number of W chromosome genes in Peking duck is more than twice that in chicken. Studying the function these female specific genes in the Peking duck will also be a very important research directions in the future.
These unique evolutionary characteristics of the duck may be related to the early differentiation of bird species, so their sex chromosomes have independent evolutionary history. Being big fans of bird genomes, and being involved in the coordinated publication of the Avian Phylogenomic Project genomes it is great to see another one take flight. With the coordinated publications today being another milestone in the study of vertebrate genomes and sex chromosome evolution.
References
Li J et al., A new duck genome reveals conserved and convergently evolved chromosome architectures of birds and mammals. GigaScience, doi:10.1093/gigascience/giaa142
Liu J et al., A new emu genome illuminates the evolution of genome configuration and nuclear architecture of avian chromosomes. Genome Res. 2021. doi:10.1101/gr.271569.120
Zhou, Y., et al. Platypus and echidna genomes reveal mammalian biology and evolution. Nature (2021). doi:10.1038/s41586-020-03039-0