Not all species are equal: Using the h-index to quantify taxonomic bias (author Q&A)
The h-index is a metric that was invented to summarise the publication output and impact of researchers. In a new GigaScience article, authors from the University of New South Wales (Australia) adopt the controversial metric for a completely different purpose: to explore systematic differences in research interest (taxonomic bias), using mammals as an example. They quantified the research interest in 7,521 species and found that many small, endangered mammals are not getting the attention they deserve.
For a Gigablog Q&A, first author Jess Tam explains the rationale of the study and what could be done to reduce taxonomic bias.
What is taxonomic bias, and why is the concept important?
“In our study, we defined taxonomic bias as uneven distributions of research across different species or taxa. Knowing which species or taxa receive more interest among scientists can help us identify gaps as well as clusters in knowledge.
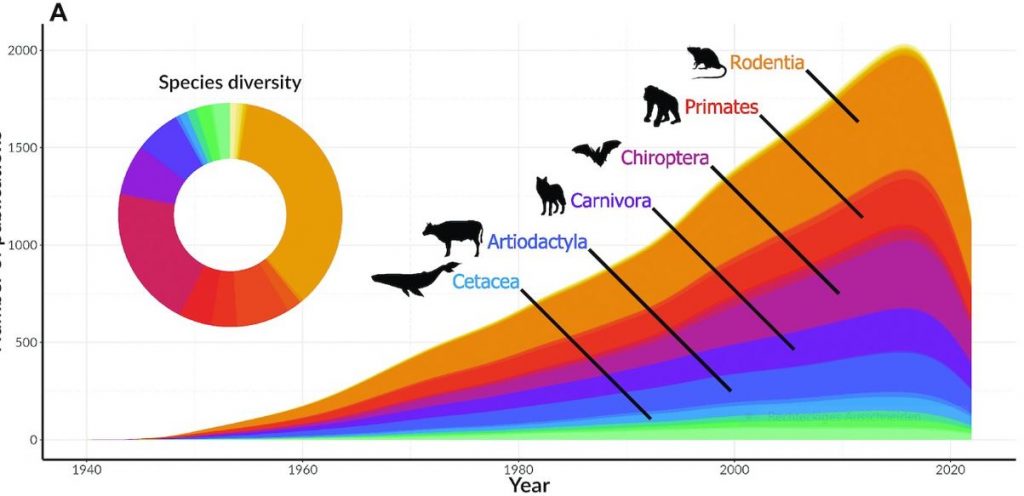
Identifying gaps in research is important because certain species with high conservation value may be left out of scientific research and conservation efforts. An example of such a species is the Cuban solenodon (Atopogale cubana). The Cuban solenodon is one of the very few species of mammals that produces venom in their saliva. While it was listed as Endangered by the IUCN Red List, we found no publications dedicated to studying this unique species. [editors’ note: The solenodon genome published by Grigorev et al in GigaScience is from a similar, but different species, Solenodon paradoxus].“

To quantify research interest in mammalian species, you’re using a metric – the h-index – that many researchers know from another context, namely, from quantifying the impact of scientific publications. What is the interpretation of an h-index, and why is it a useful metric to apply to your problem?
“The h-index is calculated by first ranking the total publications (n) from the most citations to the least citations, and then counting the number of papers with more than n citations. The h-index was originally created to summarise the research output and impact of researchers in a single number. However, this metric attracted criticism as it could encourage unfair comparisons between researchers.
In this study, we wanted to quantify the research interest each species of mammals was attracting. The h-index, we believe, is suitable here since it considers both research output (number of publications) and research attention (number of citations). There are also other metrics that could help us quantify research interest, such as the m-index. However, the m-index also considers time since the first publication as a variable, in addition to total research output and research attention, which was not part of the question in our study.”
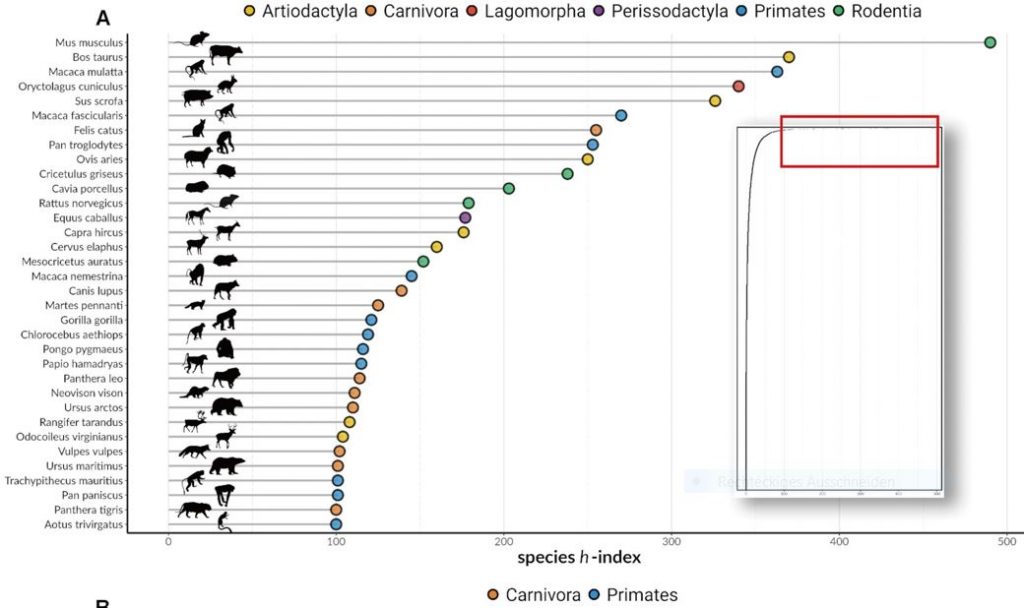
Why should we care that some mammals are less well studied than others? What do the less well studied species have in common?
“The conventional picture of a mammal is generally a big, fluffy animal. Most people associate mammals with charismatic species such as tigers, pandas, and whales (not fluffy, but really big!). This might be because charismatic animals generally receive more exposure in the media, such as books (e.g., Animal Farm by George Orwell, Moby Dick by Herman Melville), and movies (e.g., The Life of Pi, Free Willy).

While these species all occupy important niches and play important roles in their ecosystems, other species are often less visible and neglected.
Smaller mammals, such as many rodent species and small marsupials, are not very popular because of their limited coverage in the media. While some of them do appear sporadically in movies (e.g., Alvin and the Chipmunks) or in the news (e.g., when the golden bandicoot (Isoodon auratus) was reintroduced recently in Sturt National Park in Australia), charismatic species tend to attract more media attention than other species as they are more popular and familiar to the public.
A lot of small mammalian species are also critically endangered and hard to locate in the wild, which explains why both the general public and some researchers may not know of them, resulting in less academic publications and lack of fundamental knowledge.
While it is definitely a challenge (and possibly an impossible one) to study every single species on this planet, learning about the less popular species can help us appreciate the diversity of nature and understand how different species interact with each other and their environments.”
In your opinion, who is to “blame” most for the uneven research interest in mammalian species? Biased interests of scientists, funding incentives, publication bias?
“I don’t believe that taxonomic bias is necessarily a problem so wicked that it will cause the collapse of science and society. There are many personal, scientific, financial, and logistical reasons behind the decisions why scientists chose their study species. And as mentioned above, we cannot study all species. Therefore, it is no one’s fault. The government and funding bodies have varying selection criteria to follow, and researchers have different research interests and limited budgets. It is understandable that niche projects requiring more equipment and funds don’t usually succeed in securing grants. It is generally harder to convince a panel to fund a project to investigate the habitat use of a rare rodent species, than to study the population decline of the koala.

One economic solution to this problem is to study the keystone species from different habitats. Keystone species are species that affect entire ecosystems that they live in. Studying them will help us understand their interactions with other species, and broad ecosystem impacts. Keystone species are also relatively more abundant than others, such as the Tasmanian devil (Sarcophilus harrisii) in Tasmania. Another practical solution is to pick species that are easier to find and study, instead of trying to study all species. Which is likely already happening and have led to the wide gaps of knowledge in the literature today.”
Does your work provide any lessons for future “best practice”? What can researchers, funders and journals such as GigaScience do to reduce taxonomic bias?
“Some journals, though not all, have a habit of ‘gaming’ papers in order to boost their impact factors. This should be avoided if possible. Papers on charismatic vertebrate species usually attract more attention than a rare rodent that can only be found in an obscure place in a South American rainforest. While most researchers may not be interested in reading an article about a rare rodent, surely that piece of knowledge could be useful in understanding the ecology of other species or their environment. In our study, we only collected papers from peer-reviewed journals. If we had included grey literature too, we suspect that the h-index of some species might be higher than our results here.
Acquiring funding is very challenging for academics, especially those at public institutions. There are thousands of academics competing for limited pots of money. In addition to costs associated with travel expenses, equipment, and proprietary software licenses, academics must pay thousands of dollars to publish our work and attend conferences, just to share our results. The good news is that as technology is becoming more advanced and integrated with everyday life, some private technology companies have formed partnerships with different institutes to provide both technical and financial support for studying endangered species.
While I cannot provide the ‘best’ concrete solution here to bridge the gaps between existing scientific knowledge, I believe that education is key. Through constant exposure to different species, including the less popular ones, especially in children, we can help them become interested in all sorts of living species from an early age. After all, our interests can be drastically influenced by our experience and the media. Exposing ourselves to different species can help diversify our interests and hopefully fill in the gaps in scientific knowledge and conservation practice.”
Any other points you’d like to add?
“We have built an R package, specieshindex, which is now available on GitHub (https://github.com/jessicatytam/specieshindex/). We used this package here to extract citation information of papers and calculate their h-indices.”

Read the GigaScience Article:
Jessica Tam, Malgorzata Lagisz, Will Cornwell, Shinichi Nakagawa: Quantifying research interests in 7,521 mammalian species with h-index: a case study GigaScience 2022, giac074 https://doi.org/10.1093/gigascience/giac074