On the trail of the elusive Solenodon genome. Q&A with Taras Oleksyk
Researchers have sequenced the venomous Solenodon, the last survivor of a branch of mammals that appeared at the time of the dinosaurs. Here we give some behind the scenes insight with some of the authors of this prize winning project, and include a Q&A with lead author Taras Oleksyk.
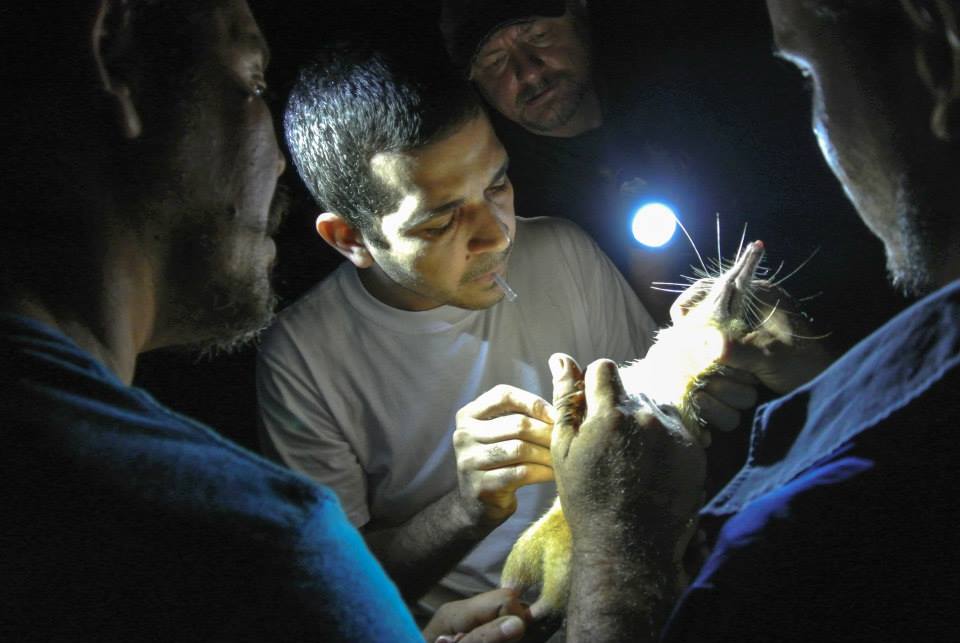
 Our latest paper presents a draft genome the venomous Hispaniolan solenodon (Solenodon paradoxus). Described by co-author Alfred Roca as “giant rat with Freddy Krueger claws“, this species is unusual not only because it is one of the very few venomous mammals, it is also the sole remaining branch of mammals that split from other insectivores at the time of the dinosaurs. The genome sequencing and analysis of this endangered animal was carried out by an international team lead by previous GigaScience author Taras Oleksyk (pictured holding one of the sequenced individuals) from the University of Puerto Rico at Mayagüez. The availability of the solenodon genome sequence allowed the researchers to answer several evolutionary questions, particularly whether the solenodon species indeed survived the meteor impact that laid waste to the dinosaurs.
Our latest paper presents a draft genome the venomous Hispaniolan solenodon (Solenodon paradoxus). Described by co-author Alfred Roca as “giant rat with Freddy Krueger claws“, this species is unusual not only because it is one of the very few venomous mammals, it is also the sole remaining branch of mammals that split from other insectivores at the time of the dinosaurs. The genome sequencing and analysis of this endangered animal was carried out by an international team lead by previous GigaScience author Taras Oleksyk (pictured holding one of the sequenced individuals) from the University of Puerto Rico at Mayagüez. The availability of the solenodon genome sequence allowed the researchers to answer several evolutionary questions, particularly whether the solenodon species indeed survived the meteor impact that laid waste to the dinosaurs.
As one of the only extant mammals that are venomous, the solenodon’s venomous saliva flows from modified salivary glands through grooves on their sharp incisors (“solenodon” derives from the Greek for “grooved tooth”). On top of the scary claws they also have several other primitive and very unusual characteristics for a mammal, including a flexible snout with a ball-and-socket joint, and oddly positioned teats, which are on their rear. While the mammalian tree of life has been heavily researched, this is the most distantly related branch to be added to the ‘genome club’. It has particular importance and implications for conservation because previous morphometric studies have suggested that southern and northern Hispaniolan solenodons may be subspecies rather than separate species.
Welcome To the club Mr. Olive, genome sequenced! @toni_gabaldon @gabaldonlab @CRGenomica https://t.co/6bgZ4BLuvM pic.twitter.com/XBnz8iaUcH
— Sketching Science (@sketchscience) June 29, 2016
Solenodon is not just genetically but also geographically isolated. Highly endangered, they remain only in a few remote corners of the Caribbean islands of Cuba and Hispaniola. Its nocturnal lifestyle makes it even more elusive and therefore less studied.
For this project, there was more than just the challenge of obtaining the organisms for blood samples, the solenodon genome proved particularly difficult to sequence. Carrying out genomics research in remote parts of the Caribbean provided a challenge, particularly in transporting high quality DNA to the lab. Due to the constraints of poor quality DNA as well as a limited budget, the commercial lab used to carry out the sequencing turned out a very low coverage per individual.
 Having already ventured into the jungle, the researchers embraced this new challenge by coming up with novel approaches to assemble the genome. First, the researchers reasoned that because the species has lived tens of millions of years in isolation it was extremely inbred and had a very homozygous genome. This lead to a potential work-around, because the five collected sets of genomic data could be pooled to increase the coverage. Despite initial doubts, this worked better than expected especially when that strategy was combined with using a string graph approach rather than the more standard de Brujn graph assembly method. String graphs incorporate more of the sequencing data than de Brujn graphs. This new technique provides a low budget alternative for genome assembly, particularly in the highly homozygous genomes of endangered species.
Having already ventured into the jungle, the researchers embraced this new challenge by coming up with novel approaches to assemble the genome. First, the researchers reasoned that because the species has lived tens of millions of years in isolation it was extremely inbred and had a very homozygous genome. This lead to a potential work-around, because the five collected sets of genomic data could be pooled to increase the coverage. Despite initial doubts, this worked better than expected especially when that strategy was combined with using a string graph approach rather than the more standard de Brujn graph assembly method. String graphs incorporate more of the sequencing data than de Brujn graphs. This new technique provides a low budget alternative for genome assembly, particularly in the highly homozygous genomes of endangered species.
The first author of the paper, Kirill Grigorev elaborates: “’For me, perhaps the most interesting part of this research was the challenge of delivering a de novo genome assembly that was suitable for comparative genomics, using an amount of sequencing data much smaller than in similar projects. With our approach, we achieved this, demonstrating that in certain cases – specifically, for species with low population heterozygosity, which is a hallmark feature of endangered species – low coverage sequencing can yield genomes comparable in quality to genomes obtained with classical approaches. Perhaps and even more important conclusion is that traditional metrics, such as the N50 statistic, do not paint the whole picture and should not be used to define the quality of an assembly alone. While our approach delivers a genome with a much smaller N50, it has much higher contiguity, which leads to more accurate gene annotation, which in turn is essential for reliable phylogenetic analyses.”.
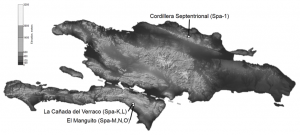
Map of where the two Hispaniolan solenodon subspecies are found.
After carrying out their assembly, the researchers had data of sufficient quality for answering many scientific questions on solenodon evolution. With regard to conservation plans the data supports that there was a subspecies split within the Hispaniolan solenodon at least 300,000 years ago, meaning the northern and southern subspecies should be treated as two separate conservation units and may therefore need independent breeding strategies.
These data also shed light on the initial speciation event for this branch, and showed that solenodons likely diverged from other extant mammals 73.6 million years ago. On the implications of this, Co-author Alfred Roca of the University of Illinois says “It is difficult to determine whether the ancestors of solenodons were already in the proto-Antilles when the asteroid that killed off the dinosaurs impacted nearby, or whether their ancestors survived on the North American mainland and later dispersed onto the island. Perhaps their Freddy Krueger-like claws allowed them to burrow their way to safety.”
 Following our tradition of author Q&As we get lead author Taras Oleksyk to provide a bit more background on the project. Taras should be well known to readers of this blog, as previous author of the Puerto Rican “peoples parrot” genome, co-organiser of the ConGen workshop we support with studentships, as well as winner of our ICG12 prize.
Following our tradition of author Q&As we get lead author Taras Oleksyk to provide a bit more background on the project. Taras should be well known to readers of this blog, as previous author of the Puerto Rican “peoples parrot” genome, co-organiser of the ConGen workshop we support with studentships, as well as winner of our ICG12 prize.
What were the challenges collecting the Solenodon?
When it comes to collecting extremely rare and secretive species in a remote locations, the challenges are many. First you have to find local contacts that will trust you and will want to work with you. Our strategy was to develop a close relationships and collaborations with our colleagues at the Instituto Tecnológico de Santo Domingo and Universidad Autónoma de Santo Domingo. After all, it is important that this project will open doors to many others to come, and we always assumed this to be the one of many projects to help research, education and conservation efforts in the Dominican Republic.
We have made many friends there, and through their contacts made crucial contacts in the local communities, and met people who could help us with the fieldwork. From the local guides Nicolas and Yimel Corona (a father and son team), we have learned much about the habits and where to find these cautious nocturnal animals. Most of the field work was done in the jungle at night, solenodons have a venomous bite, so without these local experts we stood no chance at all. Our hunting strategy was to find the trails solenodons make during daily foraging in the leaf litter, then take our position next to these trails and ambush passing solenodons at night. The hunter would have to wait for an animal to pass by for many hours in complete darkness and silence. But, ironically, an unexpected challenge comes also in the form of mountains of paperwork required to do the research on an endangered species and to transport the precious DNA samples to our laboratory in the USA.
Why was it so difficult to assemble the solenodon genome and what did these challenges teach you about trying to carry out genome projects with a low budget?
The main problem with whole genomic sequencing is transporting high quality genomic DNA to the lab. We opted to extract DNA in Dominican Republic, but during the transportation the quality has decreased, and the commercial lab turned out a very low coverage per individual (5x), which is by no means a way to assemble a genome. This was a very disheartening result and we were forced to get creative in order to rescue the project and work with the data we had. This is where Kirill Grigorev observed an unusual genome homozygosity of the genome data in our samples. This was a predicted result for a species that lived tens of millions of years in island isolation, but it also gave us a practical idea: if the five genome are practically the same, what if we could combine the samples to increase the coverage? Despite initial doubts, this worked better than expected, especially in combination with the string graph approach that seems to incorporate more of our data than the de Brujn assembly methods that currently standard in the de novo genome projects.
In essence, we have come up with a low budget alternative for a genome assembly that works fairly well in highly homozygous genomes of endangered species.
What does this data tell us about the solenodon and mammalian evolution?
Most importantly, this genome assembly allowed us to answered several important questions about solenodon evolution. First, we have confirmed the early speciation date for Solenodons, weighing on the ongoing debate on whether the solenodons have indeed survived the demise of dinosaurs after the asteroid impact in the Caribbean. Second we have confirmed the subspecies division on the island of Hispaniola, which has a direct implication for conservation. Third, we have confirmed the extreme homozygosity of this species, raising the next question – how can a species this homozygous can survive for so long? Finally, we have provided a crucial piece of the data for the following comparative studies of mammalian evolution – solenodon lineage is old and it can provide insight into the early evolution of other mammals. Solenodons have plenty of unique characters under genetic control –venom genes are just one example of these.
This research was the inaugural winner of the GigaScience prize at the International Conference on Genomics in Shenzhen at the end of last year. Presenting this work in the prize track, our international panel of judges voted it the winner of the $1000 prize and trophy. You can see a video of the talk above and also read more in the blog about the prize and its donation to the Puerto Rican students whose lives were turned upside-down by hurricane Maria. The GigaScience prize track will run again at ICG-13, and the journal will begin taking papers for it next month. Watch this space for more information.
References
Grigorev, K et al. (2018): Innovative assembly strategy contributes to understanding the evolution and conservation genetics of the endangered Solenodon paradoxus from the island of Hispaniola. GigaScience. doi: 10.1093/gigascience/giy025