Decoding The Tree of Life: Olive joins the Genome Club
Teams of scientists from Spain have published the first complete genome of the olive tree. The specimen sequenced is of the Spanish Farga variety, and is over 1,200 years old. This work will facilitate genetic improvement for production of olives and olive oil, two key products in the Mediterranean economy and diet.
Adding oil to the agricultural genomics revolution.
The olive was one of the first trees to be domesticated, roughly some 6,000 years ago. And the production of olive oil, has been documented since ~2500 BC, and was used for variety of purposes, including lamp fuel, pharmaceuticals and ritual ointments.
Despite its long history and vital economic importance, up to now the olive tree has not been part of the growing club of plant species that have their genetic code determined. Understanding its genetic instructions in this manner assists us in understanding such factors as the differences among varieties, sizes and flavour of the olives, why the trees live so long or the reasons for their adaptation to dryland farming. Other than massive difficult-to-assemble genomes such as wheat, a large proportion of all the major agricultural crops have had their genomes sequenced, published, and in many cases sequenced again using newer long-read technologies to make better quality references.
A Mediterranean emblem par excellence, the olive is of vital importance to the economy of the region. Every year, nearly three million tons of oil are produced for local consumption and export. Now a team of researchers from the Centre for Genomic Regulation (CRG) of Barcelona the Real Jardin Botánico (CSIC-RJB) and the Centro Nacional de Análisis Genómico (CNAG-CRG), has brought new insight to the genetic puzzle of the olive tree, by sequencing the complete genome of this species. The results of this have just been published with us, and pave the way to new research that will help olive trees in their development and protecting them from infections now causing major damage, such as the attacks of Olive Quick Decline Syndrome and Verticillium wilt.
“Without a doubt, it is an emblematic tree, and it is very difficult to improve plant breeding, as you have to wait at least 12 years to see what morphological characteristics it will have, and whether it is advisable to cross-breed,” says principal author of this paper Toni Gabaldón, ICREA research professor and head of the comparative genomics laboratory at the CRG. “Knowing the genetic information of the olive tree will let us contribute to the improvement of oil and olive production, of major relevance to the Spanish economy,” he adds.
Banking on open data. Private funding to support public science.
Four years ago, Gabaldón worked with Pablo Vargas, a CSIC researcher at the Real Jardín Botánico, on the presentation of scientific results of projects focused on endangered species, such as the Iberian lynx, that had been financed by Banco Santander. At that time, the bank had expressed great interest in financing scientific projects in Spain. Over the course of the presentation, Pablo Vargas proposed to Emilio Botín, the late chairman of Santander, the complete sequencing of the olive genome. A contract was signed to carry out the first complete sequencing of the olive tree’s DNA, a three-year research effort coordinated by Vargas.
Taking the olive into the genome era, this has moved the study of this ancient crop into the “big-data” arena. According to Tyler Alioto of the CNAG-CRG “this genome has generated some 1.31 billion letters, and over 1,000 GBytes of data. We are surprised because we have detected over 56,000 genes, significantly more than those detected in sequenced genomes of related plants, and twice that of the human genome.”
Despite the funding seeded from private means, to maximize its utility for scientists and olive growers, the fruits of this project have been made the public. All the sequences, transcriptomic data and results being made open source and public domain through public repositories including our GigaDB database.
Elucidating the secrets of the tree of life.
 While olive oil market has boomed on the supposed life extending benefits gained from the Mediterranean diet, the tree’s themselves may also provide some insights into the secrets of longevity. As well as the history of human migration in the Mediterranean itself. The particular specimen sequenced, named ‘Santander’, was a 1,200 year old individual of the Farga breed common in Eastern Spain (pictured here). In addition to the complete sequencing of the olive genome, researchers have also compared the DNA with other varieties such as the wild olive. They have also found the transcriptome, the genes expressed to determine what differences exist on the genetic expression level in leaves, roots and fruits at different stages of ripening.
While olive oil market has boomed on the supposed life extending benefits gained from the Mediterranean diet, the tree’s themselves may also provide some insights into the secrets of longevity. As well as the history of human migration in the Mediterranean itself. The particular specimen sequenced, named ‘Santander’, was a 1,200 year old individual of the Farga breed common in Eastern Spain (pictured here). In addition to the complete sequencing of the olive genome, researchers have also compared the DNA with other varieties such as the wild olive. They have also found the transcriptome, the genes expressed to determine what differences exist on the genetic expression level in leaves, roots and fruits at different stages of ripening.
The next step, researchers say, will be to decode the evolutionary history of this tree, which has formed part of old-world civilizations since the Bronze Age. At that time, in the eastern Mediterranean, the process of domesticating wild olive trees that led to today’s trees began. Later, selection processes in different Mediterranean countries gave rise to the nearly 1,000 varieties of trees we have today.
Knowing the evolution of olive trees from different countries will make it possible to know their origins and discover the keys that have allowed it to adapt to very diverse environmental conditions. It will also help discover the reasons behind its extraordinary longevity, as the trees can live for 3,000 to 4,000 years.
“That longevity makes the olive tree we have sequenced practically a living monument,” says Gabaldón. “Up to now, all of the individuals sequenced, from the fruit fly to the first human being analyzed, have lived for a certain time, depending on their limited life expectancy. This is the first time that the DNA of an individual over 1,000 years old, and that will probably live another 1,300 years, has been sequenced.” say Gabaldón and Vargas.
Lubricating the wheels of precision agriculture.
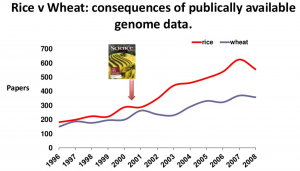 The publication of the rice genome over a decade ago has demonstrated the advantages for agricultural of having a publicly available reference genome. Rice research had a measurable boost in publications, QTLs and clones created, allowing it to overtake the previously neck-and-neck field of wheat research, that still does not have a public domain reference genome. Hopefully having an olive genome will now allow olive research and breeding to follow a similar trajectory.
The publication of the rice genome over a decade ago has demonstrated the advantages for agricultural of having a publicly available reference genome. Rice research had a measurable boost in publications, QTLs and clones created, allowing it to overtake the previously neck-and-neck field of wheat research, that still does not have a public domain reference genome. Hopefully having an olive genome will now allow olive research and breeding to follow a similar trajectory.
While most of the recent focus in genomics has been focused on using it in a clinical setting for “precision medicine”, less attention has been paid to where it is taking “precision agriculture”. This should be an equally urgent priority, with more than 1/8th of the world’s population living in extreme hunger and poverty, and the world population estimated to reach 9.6 billion by 2050. There is a huge need to create new resources to improve crop yield, reduce environmental impact, and develop crops that are of high yield and nutrition and can grow successfully in our increasingly stressed environments.
Rice has continued to show the the way ahead, with our Rice3K consortium paper presenting 3,000 rice genomes and 13TB of sequence quadrupling the amount of rice genomics data in the public domain. In the year since this was published the processed data and results has grown a further order of magnitude, with over 100TB currently available and hosted as public AWS datasets. Capitalizing on this data driven approach, breeders can now make full use of these now genetically defined strains to develop and sustain the most appropriate hybrid strains for different environments. There remains, however, one additional component to achieve this goal: information that allows researchers and breeders to directly link the genotype to the phenotype (physical traits) of these different strains. This is why on top of promoting dissemination of genetic information, we are now trying to do the same for the second part of precision agriculture: data from “big-data” phenotyping technologies such as satellites and drones, imaging techniques, timelapse, morphometrics and organ-scale phenomics. We recently launched our Plant Phenomics: Data Integration and Analyses special series, and you can see more on this on the series page and recent guest blog from the editors. Please contact us if you have important plant genotype and phenotype data you would like to disseminate in a similar manner.
