Prizes given and received, and other 2018 highlights
2018 was yet another eventful year at GigaScience. Before most of the editors and data curators take a few days of well-earned break from handling Giga-papers and Giga-data, it’s time to look back over this year’s highlights – of which there are many. Prizes were handed out and received, we travelled to conferences all over the world, and, most importantly, we published outstanding research and data.

All I want for Christmas is Tegu!
Gigascience Prize Track at ICG
As in the previous year, we invited authors of cutting edge, unpublished research to have their submission to GigaScience considered for our prize track at BGI’s ICG (International Conference on Genomics) conference in Shenzhen, which was held in October.
We invited the authors of submissions that were accepted for the series to present their work at the conference, with complimentary registration and compensation for travel.
This year’s winners convinced the jury also because they showed the value of mining existing datasets for hidden gold: Titus Brown and his lab presented an automated pipeline to re-analyze a massive marine microbial dataset from the Microbial Transcriptome Sequencing Project (MMETSP). First author Lisa Johnson travelled to ICG to present the work. To learn more about the study, follow this link to an “Author Q&A” on GigaBlog. In addition, the authors also published a commentary, proving further background on the importance of reproducible and re-usable pipelines.
The associated publications of the other winners of the prize track are also feeding through to the journal and series page, and you can read Q&A’s with some of the other winners such as Anil Thanki, and see their talks in our GigaTV youtube channel.
We won an award!
 At ICG, we handed out prizes, but in 2018 we were also at the receiving end of recognition for our efforts: GigaScience, together with our publisher OUP, won the PROSE Award for Innovation in Journal Publishing in the multidisciplinary category, by the Association of American publishers (AAP). GigaScience and its associated database GigaDB, in cooperation with services such as Protocols.io and Code Ocean, is moving beyond the traditional research paper, for the sake of reproducibility and open data sharing. We are doing this because we are convinced this is how research should be shared in the 21st century, and we’re happy that the publishing professionals at AAP recognized our efforts as a prize-worthy innovation. You can see a preprint covering the latest technical advances and updates to our GigaDB platform in bioRxiv here.
At ICG, we handed out prizes, but in 2018 we were also at the receiving end of recognition for our efforts: GigaScience, together with our publisher OUP, won the PROSE Award for Innovation in Journal Publishing in the multidisciplinary category, by the Association of American publishers (AAP). GigaScience and its associated database GigaDB, in cooperation with services such as Protocols.io and Code Ocean, is moving beyond the traditional research paper, for the sake of reproducibility and open data sharing. We are doing this because we are convinced this is how research should be shared in the 21st century, and we’re happy that the publishing professionals at AAP recognized our efforts as a prize-worthy innovation. You can see a preprint covering the latest technical advances and updates to our GigaDB platform in bioRxiv here.
ISMB
As we launched at the ISMB (Intelligent Systems for Molecular Biology) conference in 2012, every year since we’ve held our birthday celebrations at the meeting. This year it was off to Chicago, where thanks to generous sponsorship from OUP we hosted a 6th birthday party at the Watershed bar, and much craft beer and GigaPanda cake was consumed by all. Our lead Biocurator Chris Hunter also presented a poster on the most recent GigaDB development. You can see the pictures in our facebook gallery, and look forward to our 7th birthday in Basel next year.
DNA Day: Genomics in the jungle

Aaron Pomeratz in his natural environment – sequencing DNA in the jungle.
April 25th is DNA day – commemorating the day when, in 1953, the structure of DNA was announced. Here in GigaBlog, we celebrated the day by posting an author Q&A with Aaron Pomerantz, talking about his experience with DNA sequencing in the field. Rather than shipping DNA from the field to the lab, he brought a portable DNA sequencer to the jungle and analyzed his samples right on the spot. Aaron explained why this is an exciting new option: “It is a tool to quickly obtain genetic information that can be used to identify species/populations, establish geographic distributions, aid with in situ conservation actions, ex situ breeding programs, and promote conservation law efforts.”
The resulting article and data were published in GigaScience and GigaDB, respectively.
Democratising Data
 In November, Gigascience organized a session on ”Democratising Data Publishing: A Global Perspective” at the International Data Week conference in Gaborone, Botswana. As our executive editor Scott Edmunds reported here on GigaBlog, “it was refreshing to reach a critical mass of diverse voices that were not the usual subjects from the well funded European and North American institutions, and everyone who made it to Botswana felt it was a historic and eye opening meeting because of it.“ Speakers at the track included GigaScience Data Scientist Chris Armit (pictured), who focused on many of our agricultural data projects (slides here) and our Editorial Board Member Susanna Sansone, who talked talked about FAIRsharing (see slides), among others.
In November, Gigascience organized a session on ”Democratising Data Publishing: A Global Perspective” at the International Data Week conference in Gaborone, Botswana. As our executive editor Scott Edmunds reported here on GigaBlog, “it was refreshing to reach a critical mass of diverse voices that were not the usual subjects from the well funded European and North American institutions, and everyone who made it to Botswana felt it was a historic and eye opening meeting because of it.“ Speakers at the track included GigaScience Data Scientist Chris Armit (pictured), who focused on many of our agricultural data projects (slides here) and our Editorial Board Member Susanna Sansone, who talked talked about FAIRsharing (see slides), among others.
The African Orphan Crops Consortium bears its first fruits
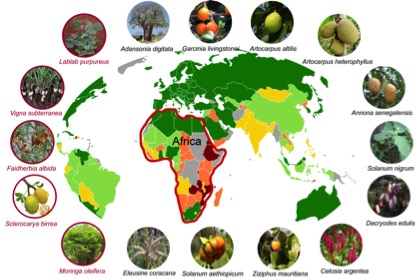
The African Orphan Crops Consortium (AOCC) is an international public–private partnership, co-led by African Union’s New Partnership for Africa’s Development (AU-NEPAD Agency), World Agroforestry Centre (ICRAF), BGI, and University of California, Davis. AOCC is planning to sequence 101 plants that are so far neglected by scientists, but contribute to traditional African food supplies. In December we published the first fruits of this effort: A report on the sequencing, assembly, and annotation of five genomes of underutilized plants. Of course, the articles are open access and all data is released without restrictions via NCBI and GigaDB.
There is no shortage of other outstanding examples of Big Data science published in Gigascience during the course of 2018. It’s hard to pick just a few highlights. Here are some of our personal favorites, with apologies to the authors of the many excellent articles that we can’t cover in this blog:
The Giardia proteome
Kevin Tyler and his team at UEA’s Norwich Medical School used state-of-the-art proteomics to study how the Giardia parasite can cause severe symptoms in some patients. Kevin explained here on GigaBlog why the work matters: “When we talk about infectious diseases in the developed world, there is a focus on viral and bacterial infections. Parasites are sometimes left out of the conversation because in the world’s wealthier countries they cause few hospital admissions. World-wide however, protozoan parasites cause the highest burden of disease”.
To better understand how the Giardia parasite causes infection, the authors took an in-depth look at the proteome:
“Our approach was to identify and quantify all the proteins in the trophozoites [activated, feeding stages] that current detection technology permits and to compare the abundance of these with what we found in the media in which we grew them, to look for enrichment in the media as a signature of actual secretion.”
Genome of a venomous mammal, the Solenodon

Over the years, we published genomes of lots of bizarre organisms, but the Solenodon is in a league of its own in this respect: not only is it venomous (very unusual for a mammal), it also displays a number of other special features, such as a flexible snout with a ball-and-socket joint and teats which are placed at the rear end of the animal. The research by Taras Oleksyk and his team was the inaugural winner of the GigaScience prize at the International Conference on Genomics in Shenzhen at the end of 2017.
Genome scale model of a superbug
Antibiotics expert Jian Li and his team at Monash University published a genome-scale metabolic model of the pathogen P. aeruginosa, which is the most complete metabolic reconstruction of this particular pathogen to date. It includes 3022 metabolites and 4265 reactions and will help scientists to explore bacterial responses to antibiotics more easily and provides a short-cut to time-consuming and costly experimental work (see also our blog post).
Genome of the tegu
The quality of genome assemblies published in GigaScience has been rising substantially recently, and most of the genome data notes we consider these days present extremely high (“platinum”) quality, chromosome scale assemblies, using a range of techniques to improve and polish the result. A prime example for this development is the genome of the tegu, a charismatic lizard from South America that has taken a step towards being full-blown warm-blooded (and it’s also cherished as an an exotic pet). The authors of the GigaScience paper, Michael Hiller and his team, used a range of techniques, also including optical mapping, and rewarded themselves with the most complete assembly of any reptile genome so far.
For GigaBlog, the authors explained some of the challenges they had to overcome: “We had to integrate different sequencing and scaffold technologies, which turned out to be difficult since existing tools often don’t work as expected, especially if the input data differs from what the tool was initially developed for. It took numerous rounds of trial and error to find a combination of tools that substantially improved the existing short-read based genome assembly using long-read sequencing data.”
A micro X-ray computed tomography dataset of South African hermit crabs
With our innovative publishing model, combining research articles with a data repository, GigaScience is the place to go if you wish to share large imaging datasets as part of your published research. A beautiful example for this is the Data Note by Jannes Landschoff and colleagues, presenting computer tomography 3D-reconstructions of five South African hermit crab species. The Data Note describes the methods of image acquisition and provides context for the work, and our repository GigaDB hosts the virtual 3D specimen and 3D printable versions. We now have sketchfab and thingiverse accounts to allow interaction with these 3D models, and a platform for the 3D printing community to discuss and share this work.
We look forward to 2019 and wish all our readers, authors, reviewers and editorial board members a Happy New Year!